Marika Kapsimali and Steve Wilson
Original paper reference
MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system
Our genome contains the instructions, in the form of genes, for making tens of thousands of proteins that are the building blocks and machinery of all the cells in our body. The genome is composed of DNA and each gene is a stretch of DNA that encodes a matching RNA that is then used as a template for building proteins. In recent years, it has become evident that not all RNAs that are encoded by the genome are used to make proteins. There are lots of very small RNA molecules called microRNAs or miRNAs that are thought to regulate whether other, larger, RNAs are made into proteins or are degraded. In this study, we aimed to examine the expression of lots of miRNAs in the developing and mature brain to find out which groups of neural cells might be using each of the different miRNAs. We reasoned that a detailed analysis of miRNA expression in different brain areas would be a useful resource for studies in the future that try to address the functions of the various miRNAs
Previous work in Ronald Plasterk's lab (Science 2005 309:310-1.) had identified a considerable number of miRNAs expressed in the zebrafish brain. For our study, we chose 38 that are conserved in vertebrates and have brain specific expression. A method called in situ hybridization was already established to detect their expression (Nat Methods. 2006 3:27-9.). The principle is to incubate cells with a labelled modified nucleic acid (Locked Nucleic Acid, LNA) that is complementary in sequence to the mature miRNA - if the miRNA is present in the cell, the complimentary LNA will bind to it and as the LNA can be visualised (the blue staining in the pictures below), one can determine which cells express the miRNA (the blue ones) and the ones that don't (pink in the pictures). Once the data on miRNA expression were gathered, we identified the brain areas where they expressed with the help of available neuroanatomical atlases (zfin.org).
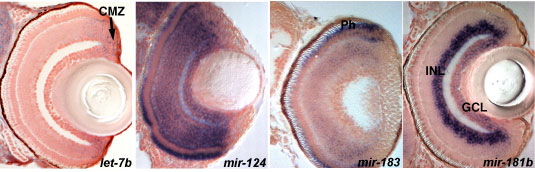
Images of sections through the eyes showing a variety of expression profiles for different miRNAs. miRNA expression is detected by in situ hybridization (blue) and the sections are counterstained with nuclear red stain to visualize the cell nuclei. let-7b is expressed in proliferating cells (arrow, CMZ-ciliary marginal zone), mir-124 in the all differentiated retina cells, mir-183 in photoreceptors (Ph) and mir-181b in a subset of differentiating cells (INL-inner and GCL-ganglion cell layers).
The results show that miRNAs have a wide variety of different expression profiles in neural cells including: expression in proliferative cells and/or their differentiated progeny, regionally restricted expression, cell-type specific expression, constitutive expression in mature neurons. This suggests several modes of miRNA action in the neural cells. For example, cell-type specific miRNAs may modulate the spatial and/or temporal regulation of target mRNA translation within mature neuronal cells. Overall this survey provides an important resource for future functional studies of miRNAs in the brain.
Other links
Faculty of 1000 assessment
Scientific reviews
Bartel DP. Cell 2009 136:215-33. MicroRNAs: target recognition and regulatory functions.
Asli NS and colleagues. Curr Mol Med. 2008 8:698-710. MicroRNAs in organogenesis and disease.
Kosik KS. Nat Rev Neurosci. 2006 7:911-20.The neuronal microRNA system.