A summary of the article “Compensatory growth renders Tcf7l1a dispensable for eye formation despite its requirement in eye field specification” Elife. 2019 Feb 19;8. pii: e40093. doi: 10.7554/eLife.40093.
Although some genetic mutations lead to congenital abnormalities and/or disease, DNA sequencing projects have shown that compromised function of many genes has no obvious effect. Indeed, people that carry such mutations live an apparently normal life that is not impacted by altered gene function. This suggests that genetic pathways and developmental processes are robust and able to cope when challenged by altered gene function. An important goal in biomedicine is to identify the compensatory mechanisms that enable such mutations to be carried without consequence
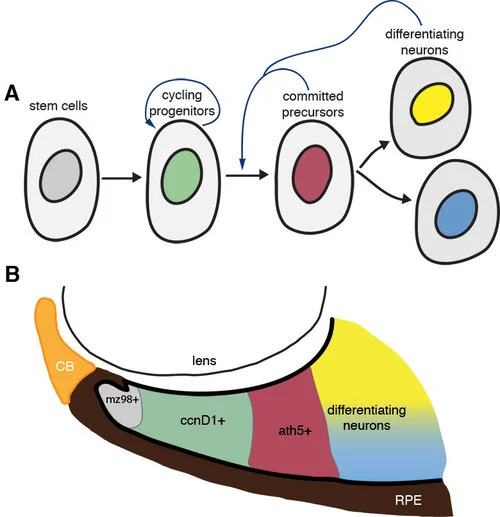
Zebrafish embryos are well suited to explore the link between genetic mutations (genotype) and developmental outcome (phenotype). In a paper recently published in eLife (elifesciences.org/articles/40093), we studied zebrafish carrying a mutation in a gene called tcf7l1a that encodes a protein functioning in the Wnt pathway, one of the most critical pathways by which cells signal to each other during development. The Wnt pathway is particularly important for development of the eyes. Zebrafish embryos with the tcf7l1a mutation initially have tiny forming eyes, but within a day or two, these eyes undergo compensatory growth to reach a size comparable to that in unaffected individuals. We found that the smaller eyes delayed the onset of differentiation, enabling them to continue growing until they reached the right size. How does the eye know what the right size is? We propose that there is a mechanism by which eye size is intrinsically assessed and this informs retinal cells when to stop dividing and start differentiating into neurons. This mechanism may also explain how it is that left and right eyes develop independently and yet still grow to the same size.
Although compensatory growth could restore eye size when tcf7l1a function was absent, we speculated that developing eyes in these mutants may be very sensitive to the effect of additional genetic mutations. Indeed, we found additional interacting mutations that completely impaired eye development such that no eyes were formed, and others in which the developing eyes were unable to compensate their growth and remained small.
These results contribute to a better understanding of the origin of developmental eye pathologies such as microphthalmia and anophthalmia, in which people are born with small eyes or no eyes at all.
Read the digest of this paper on Elife or treat yourself to the full text here.
Key Publications
Young RM, Hawkins TA, Cavodeassi F, Stickney HL, Schwarz Q, Lawrence LM, Wierzbicki C, Cheng BY, Luo J, Ambrosio EM, Klosner A, Sealy IM, Rowell J, Trivedi CA, Bianco IH, Allende ML, Busch-Nentwich EM, Gestri G, Wilson SW.
Compensatory growth renders Tcf7l1a dispensable for eye formation despite its requirement in eye field specification.
Elife. 2019 Feb 19;8. pii: e40093. doi: 10.7554/eLife.40093.
Sizing the eyes-a study of the mechanisms by which the developing eye copes with deleterious genetic mutations