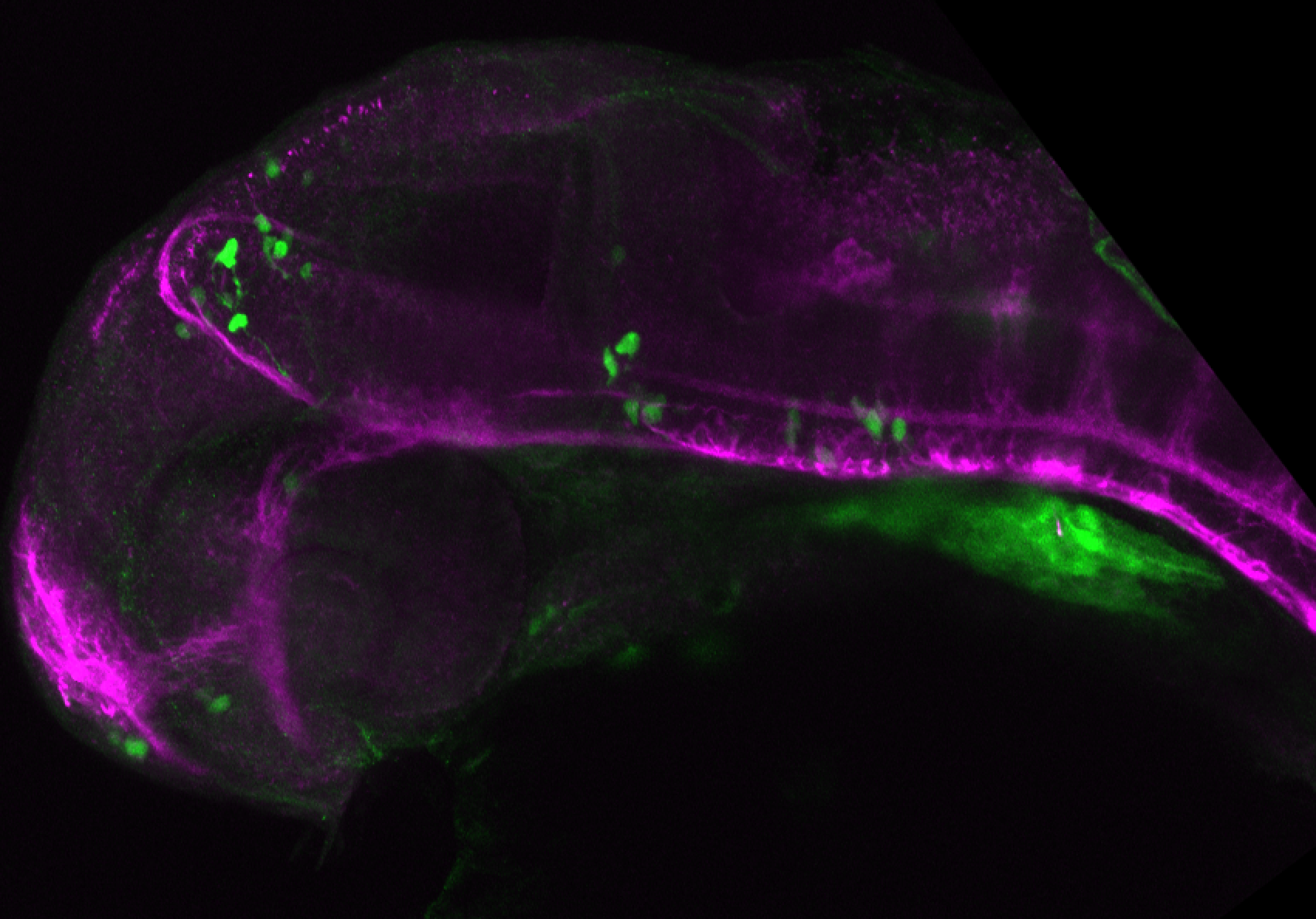
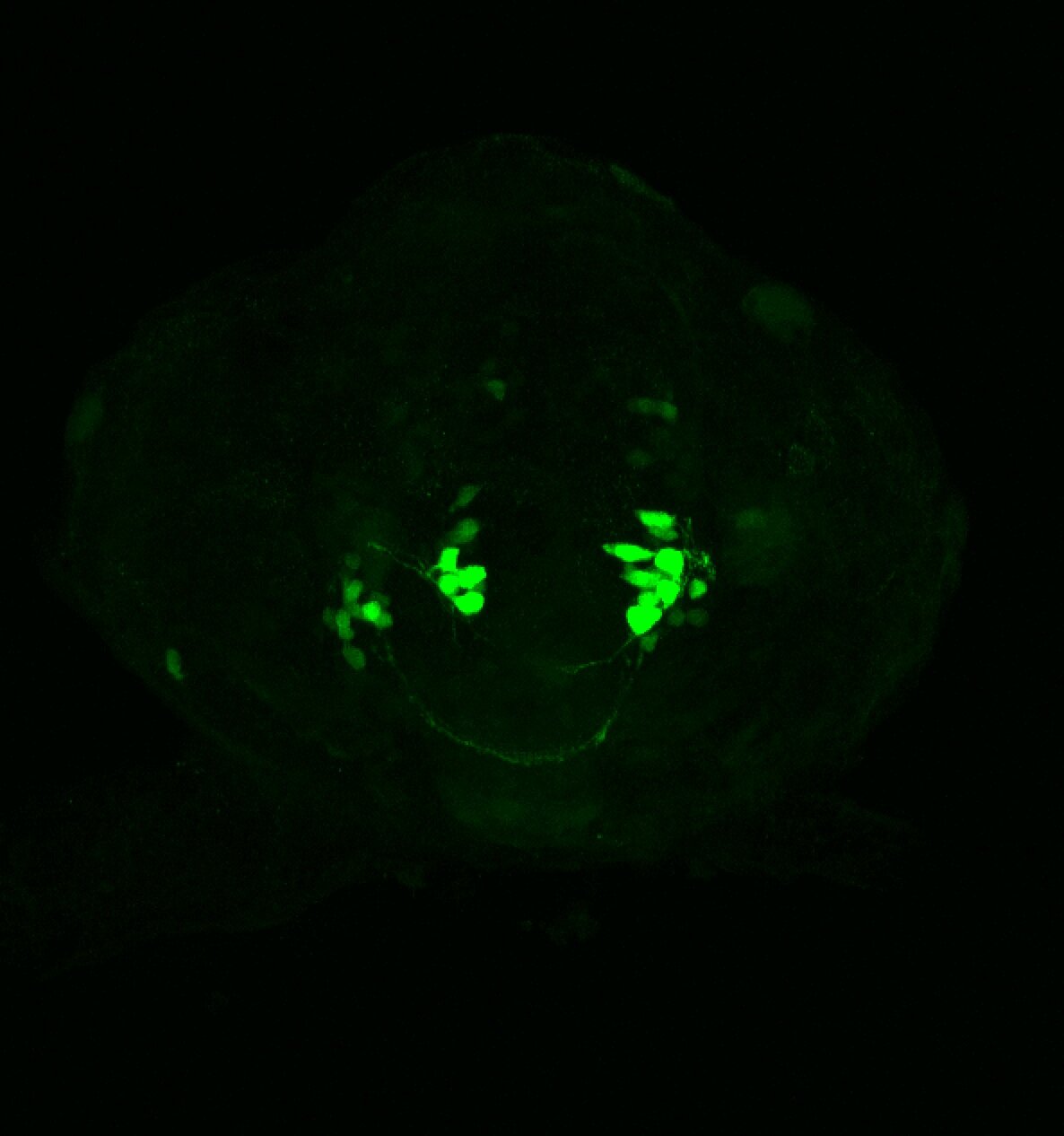
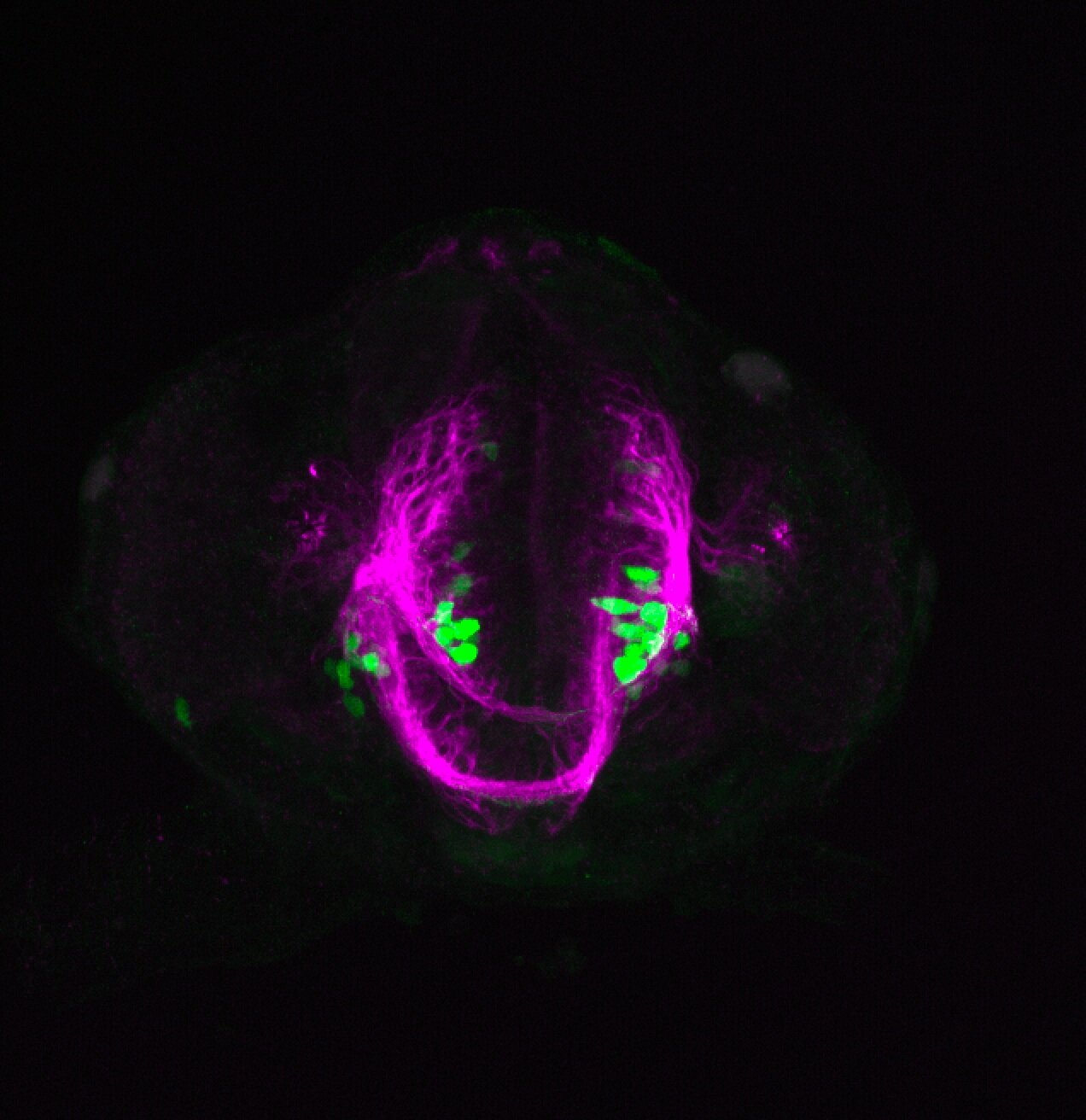
About
construct:
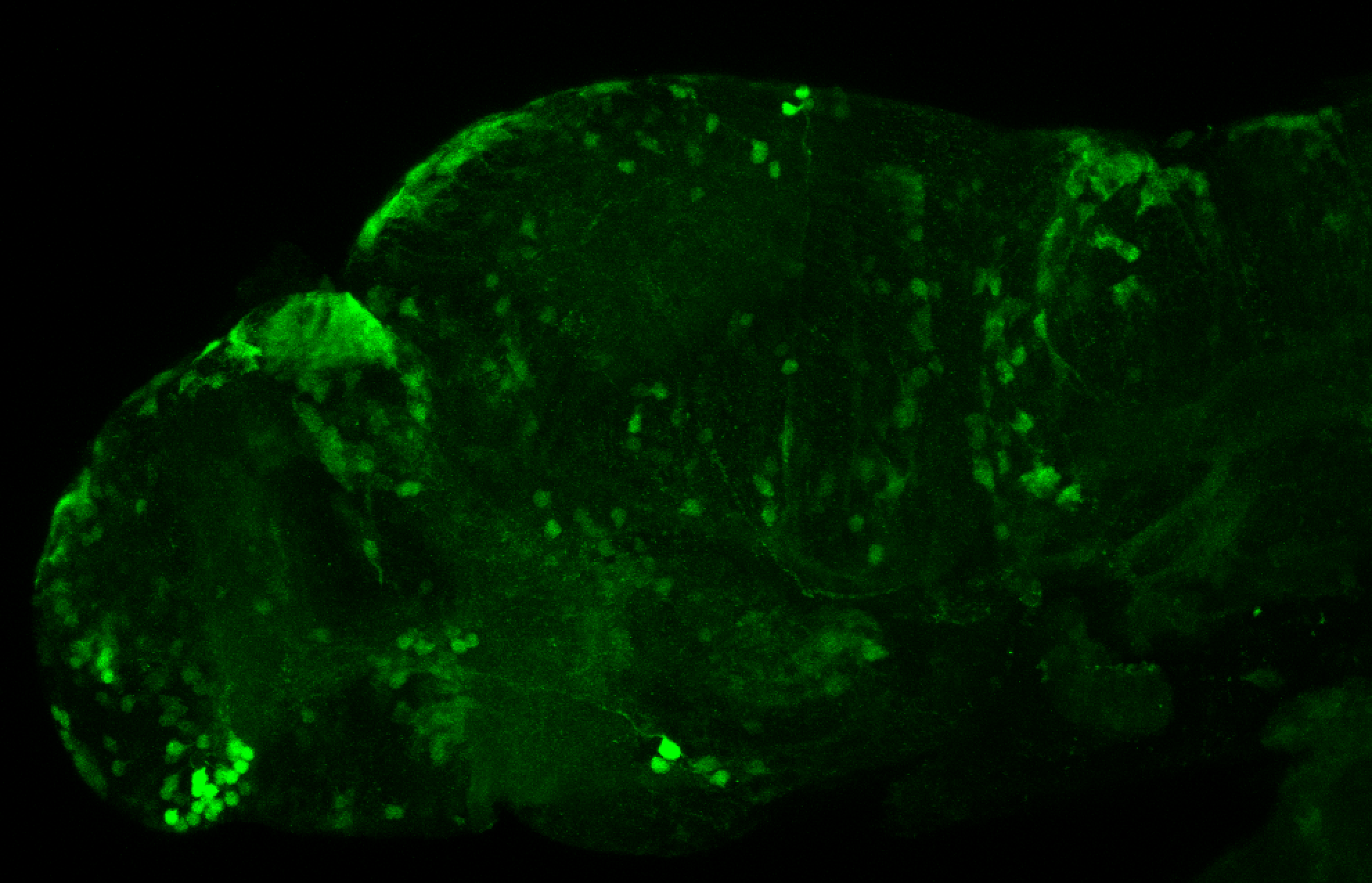
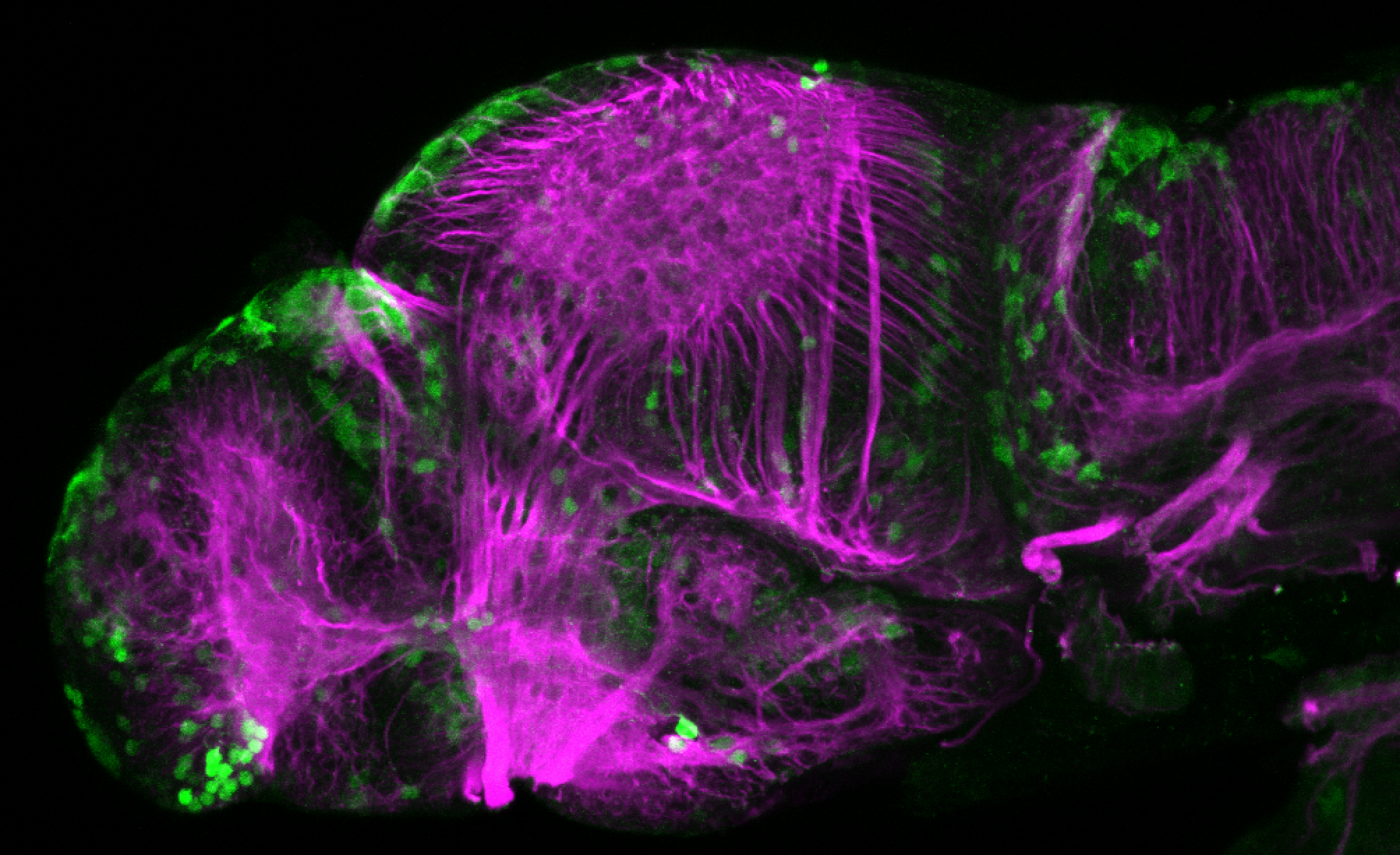
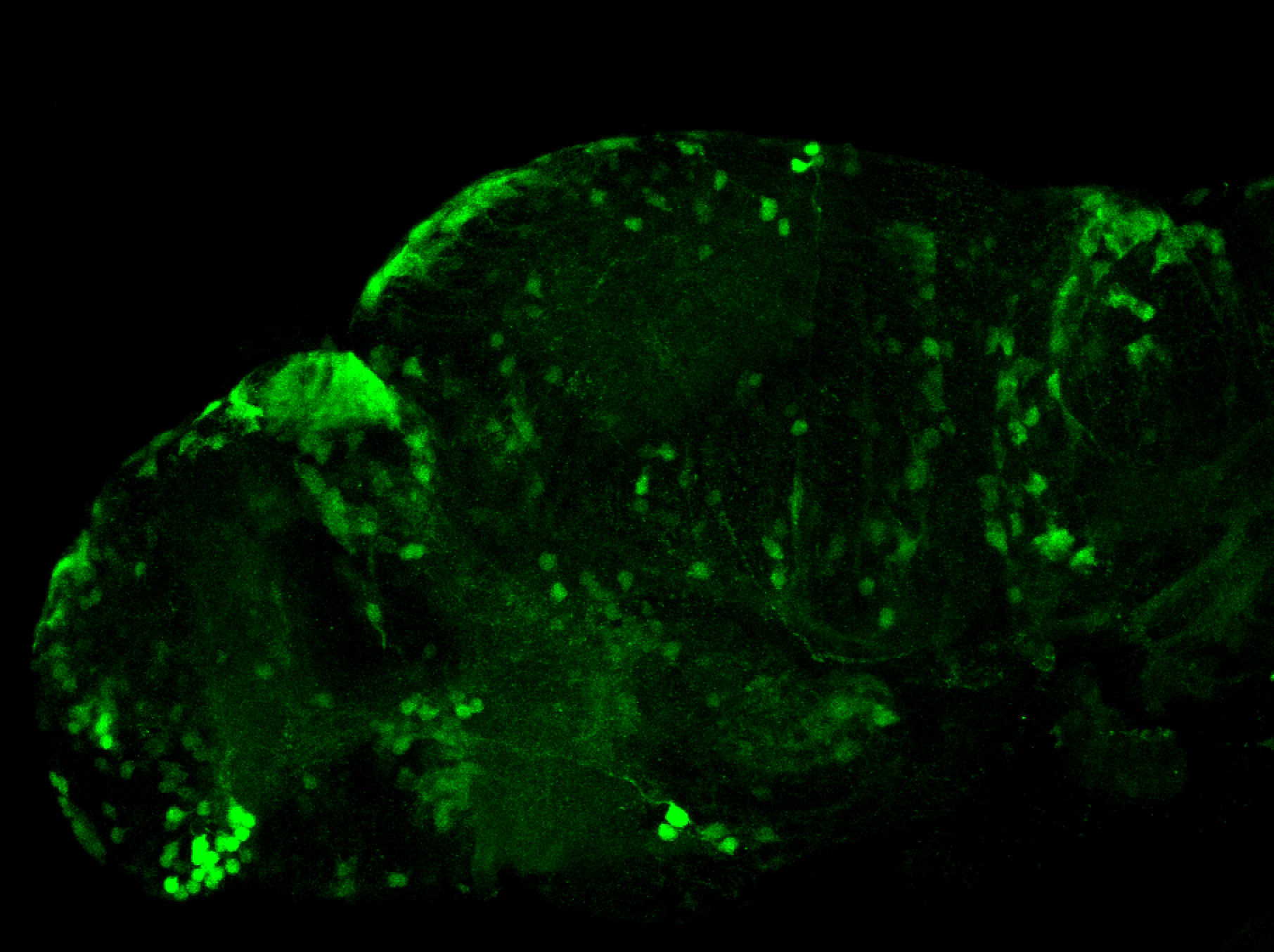
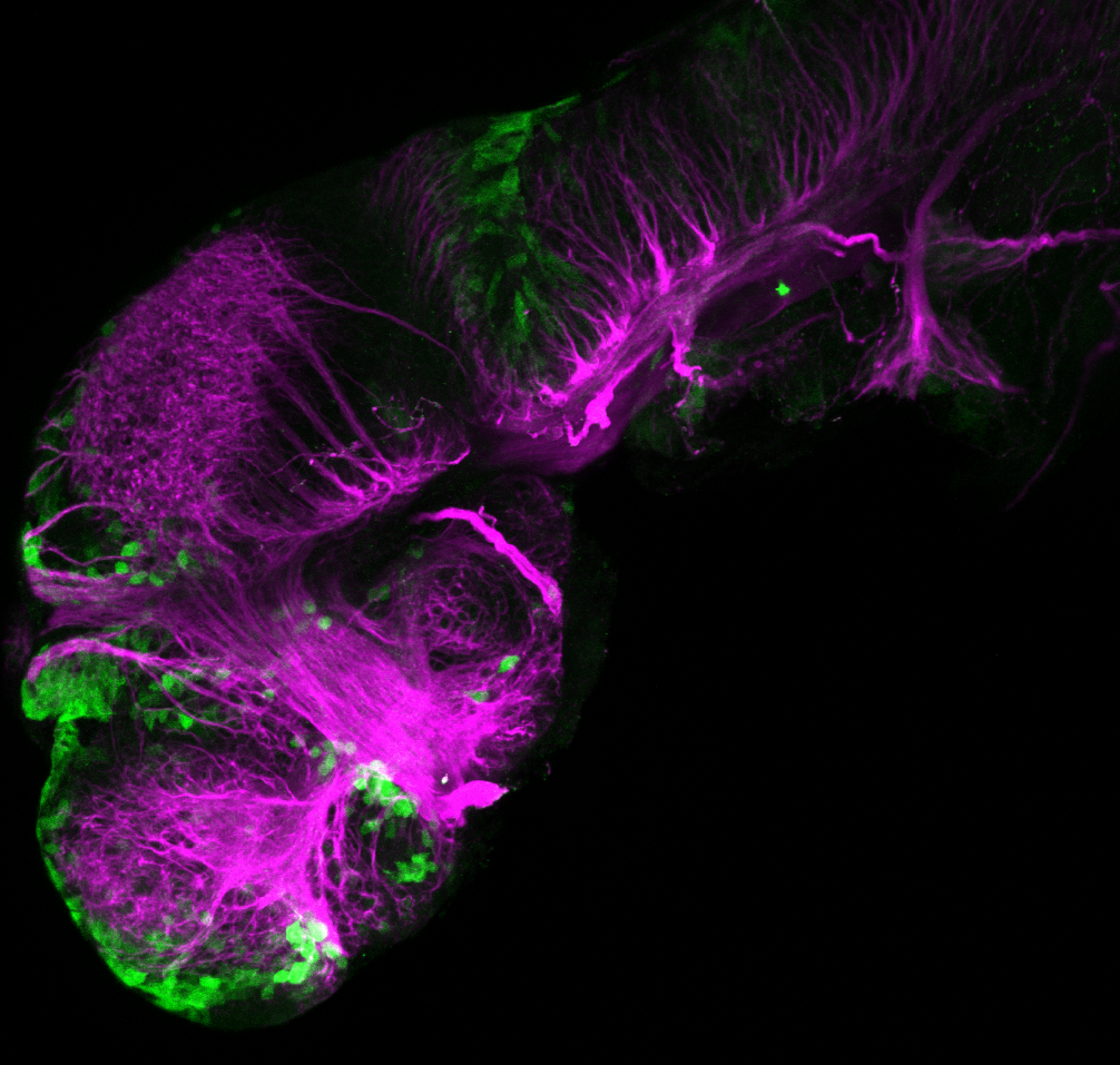
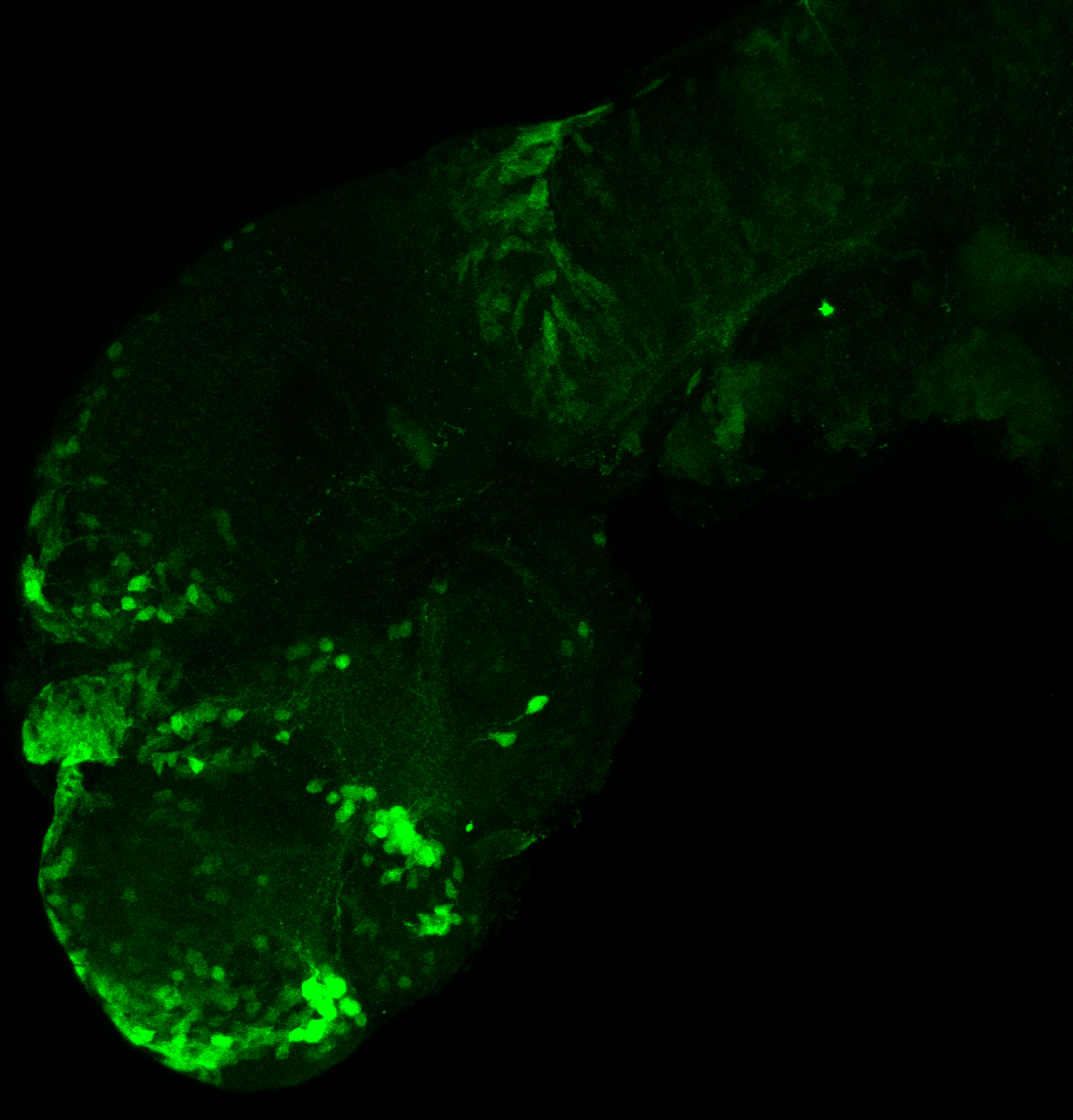
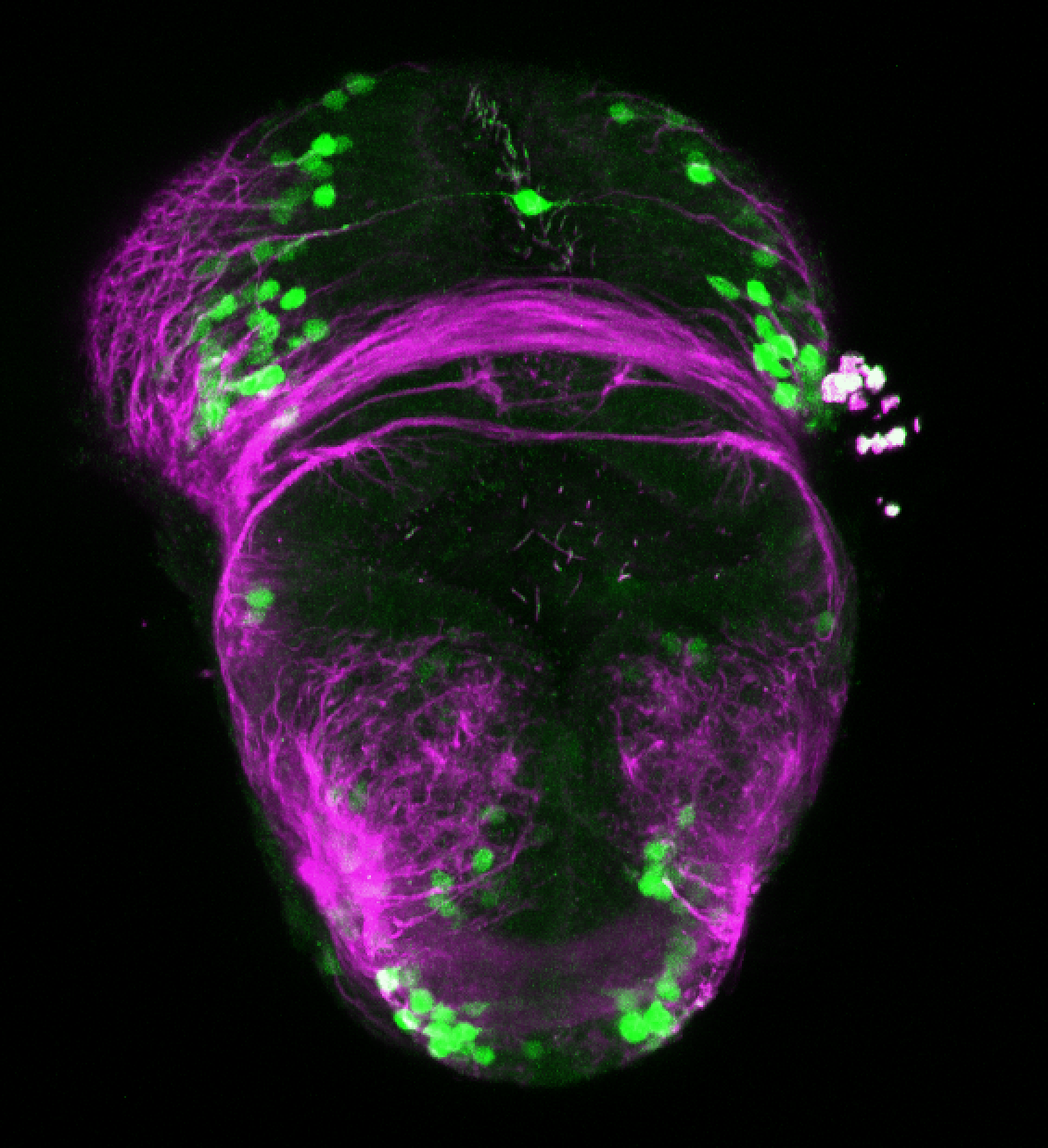
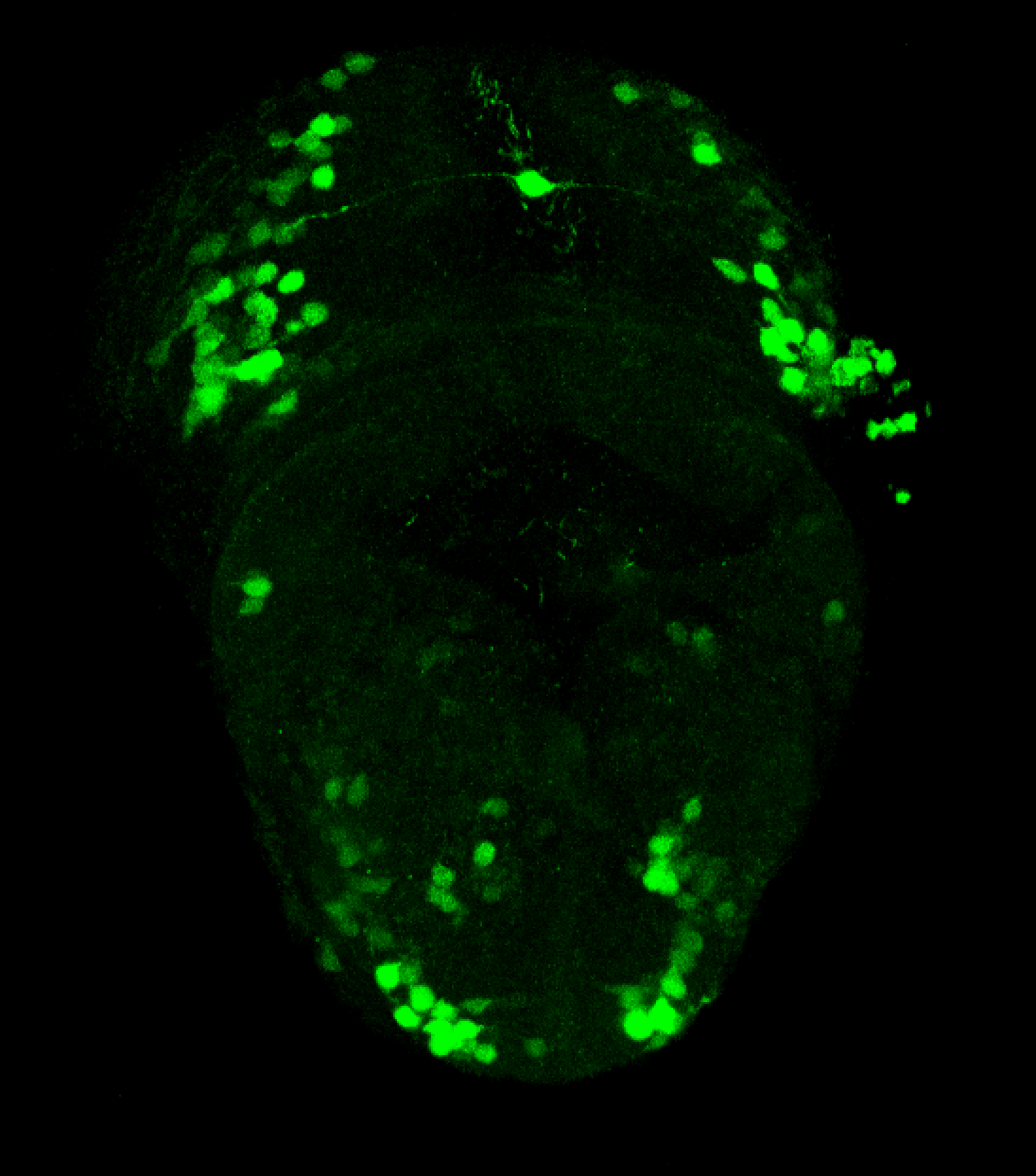
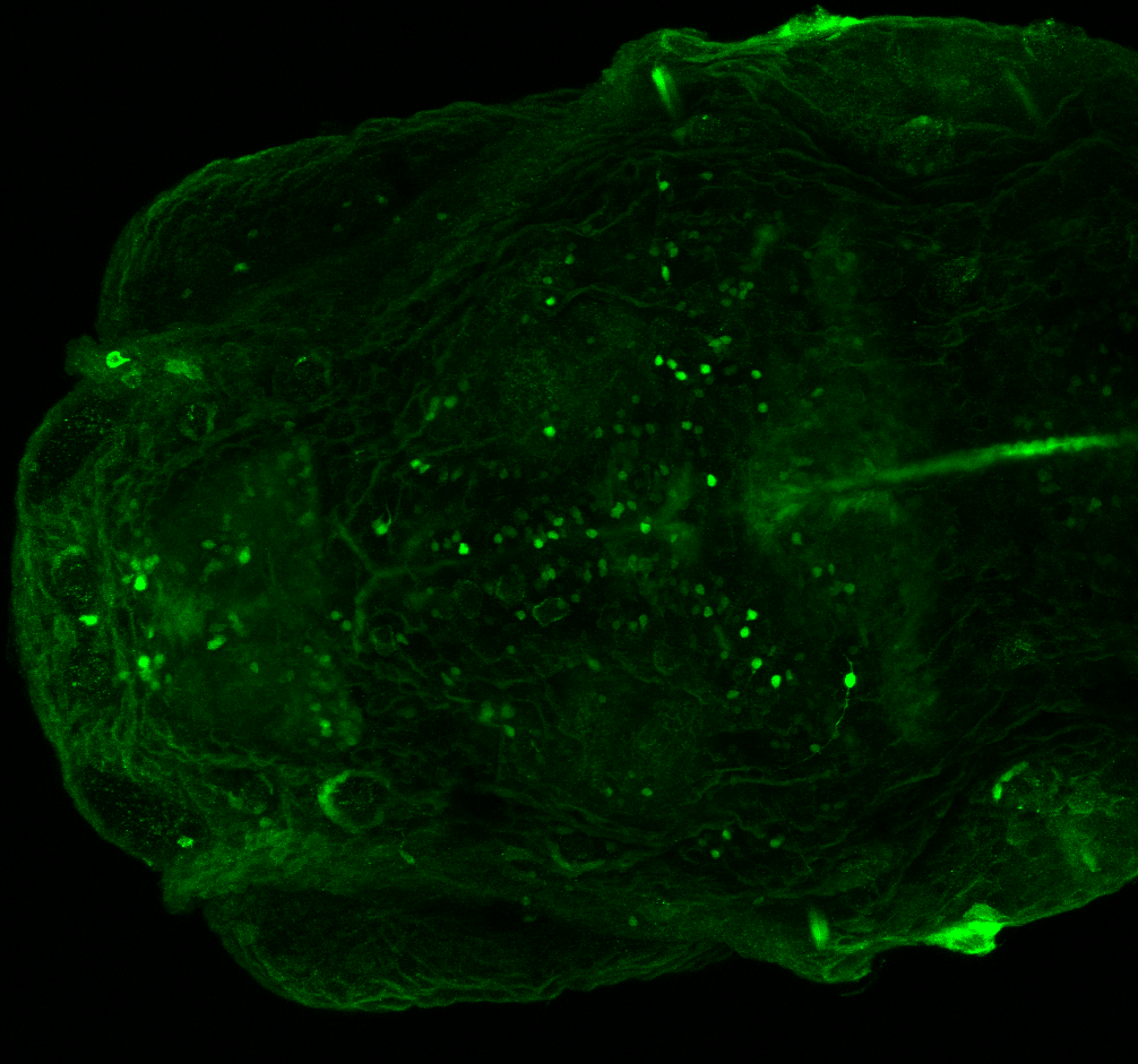
Tg(-8.0cldnb:LY-EGFP)
This transgenic line was created by the Gilmore lab “ Eight kilobases of sequence directly upstream of the Claudin B start codon were amplified from BAC zK241F11 by using the Expand Long Template PCR System (Roche). The resultant fragment was cloned into a vector containing lynEGFPpA (Koster and Fraser, 2001) flanked by sites for I-SceI, and the resultant construct was injected into one-cell zebrafish embryos by following the meganuclease transgenesis protocol (Thermes et al., 2002).”(Haas & Gilmore, 2006).
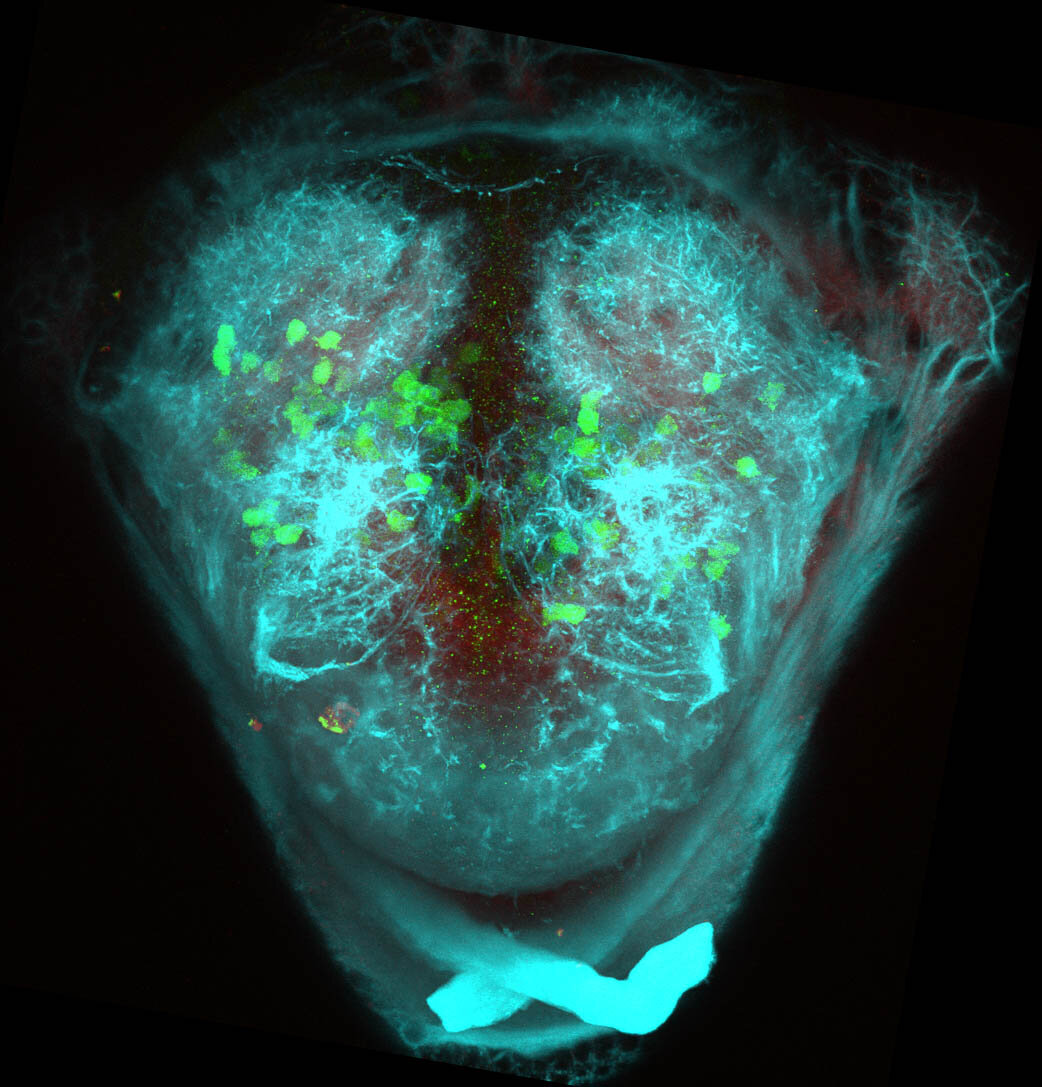
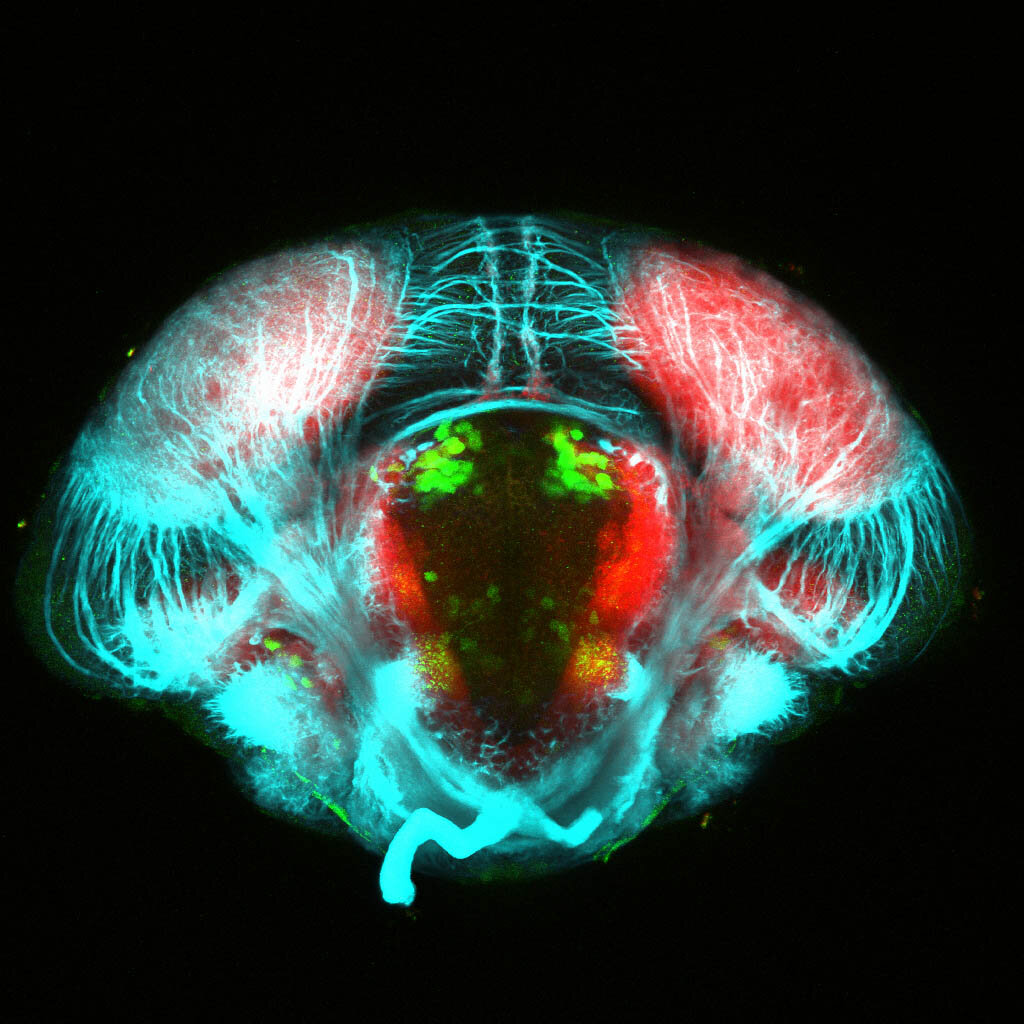
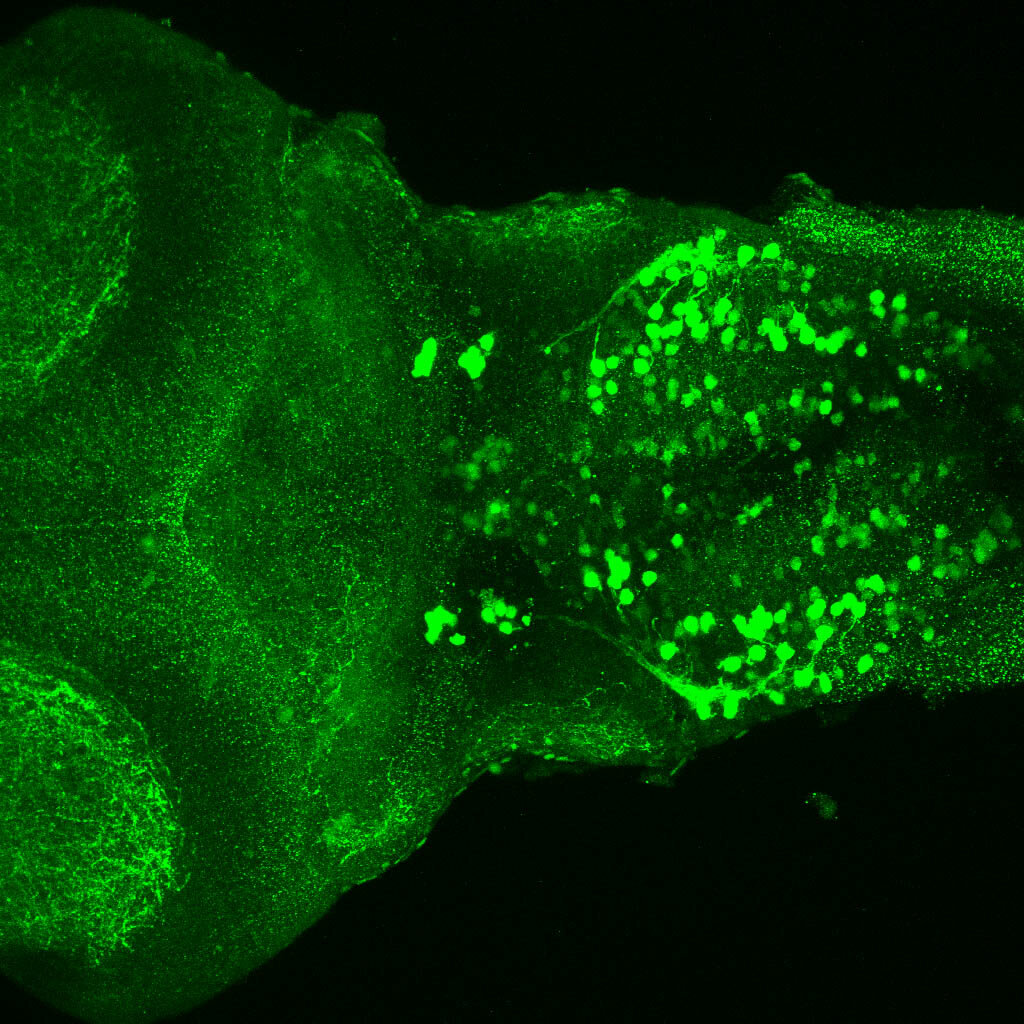
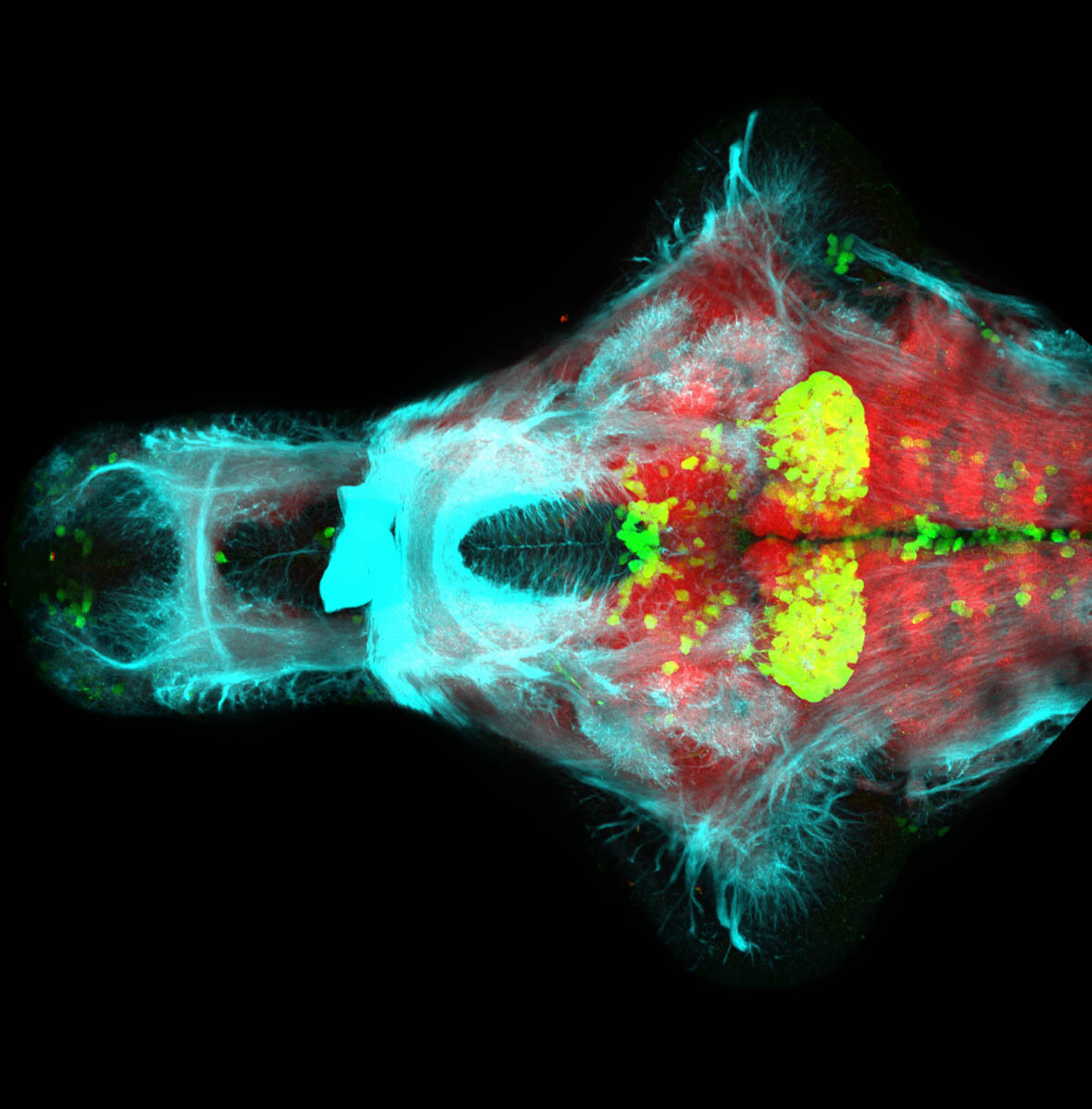
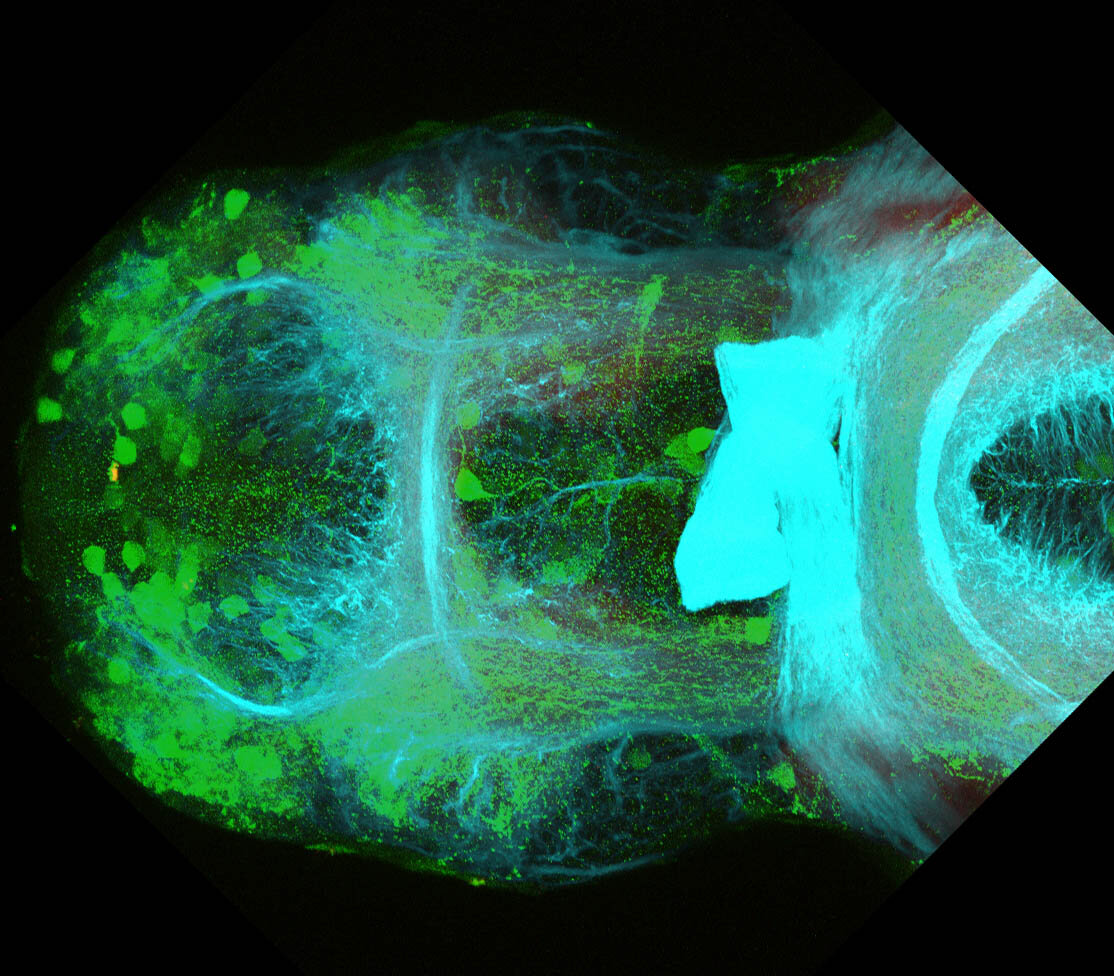
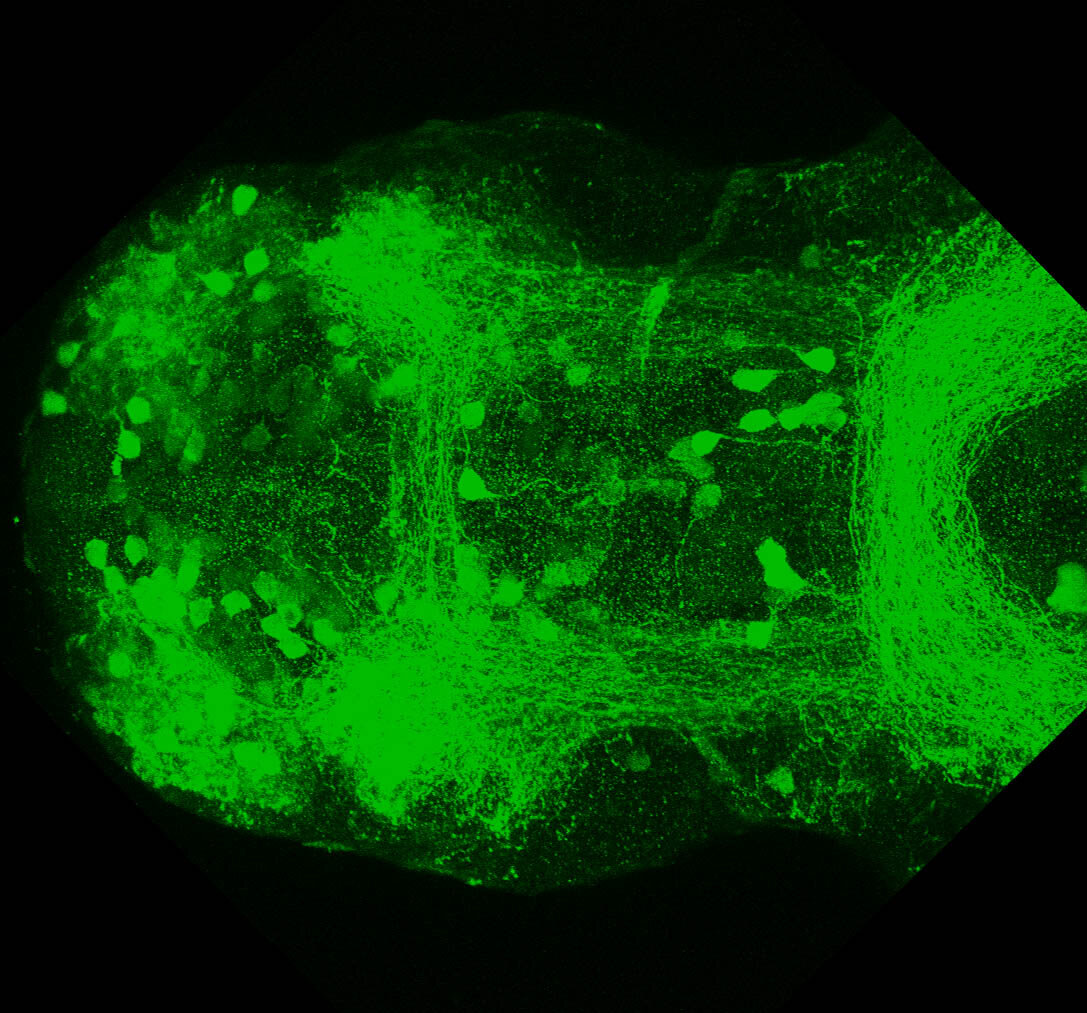
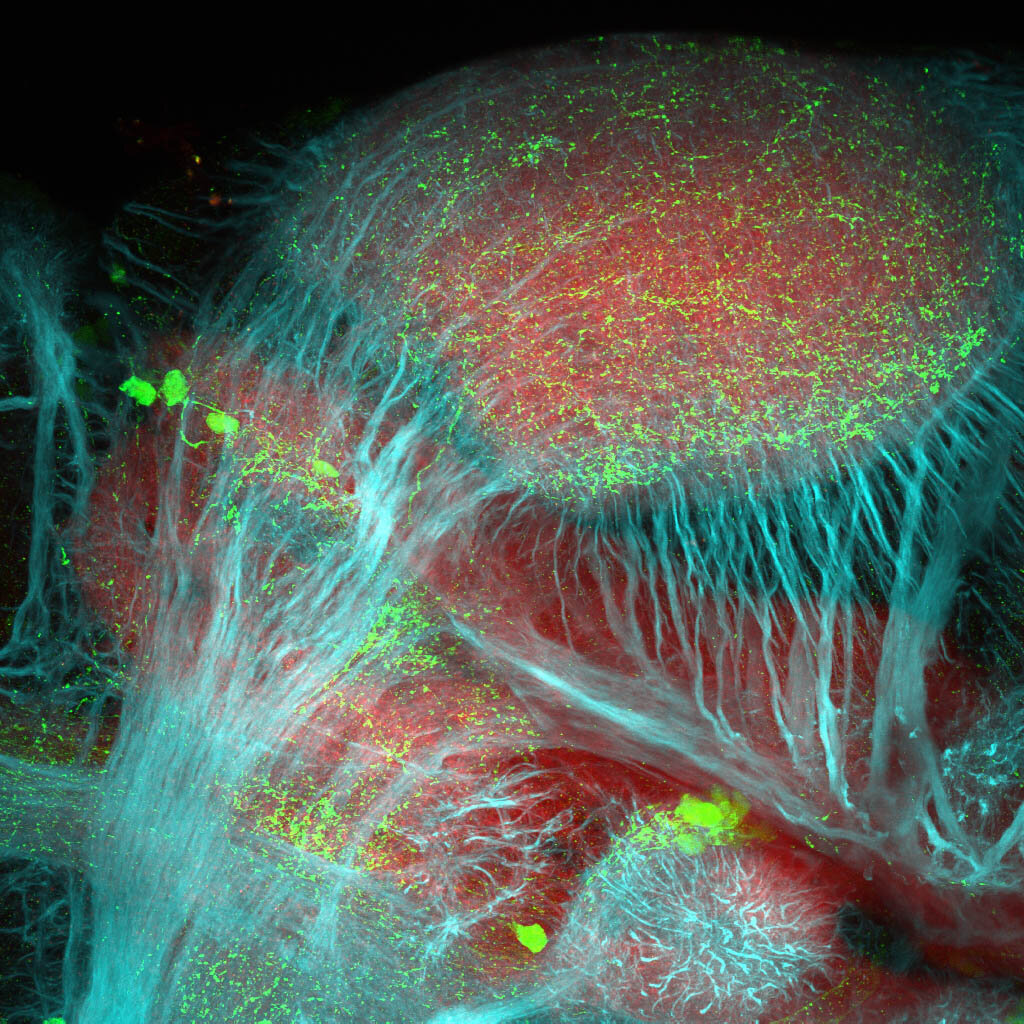
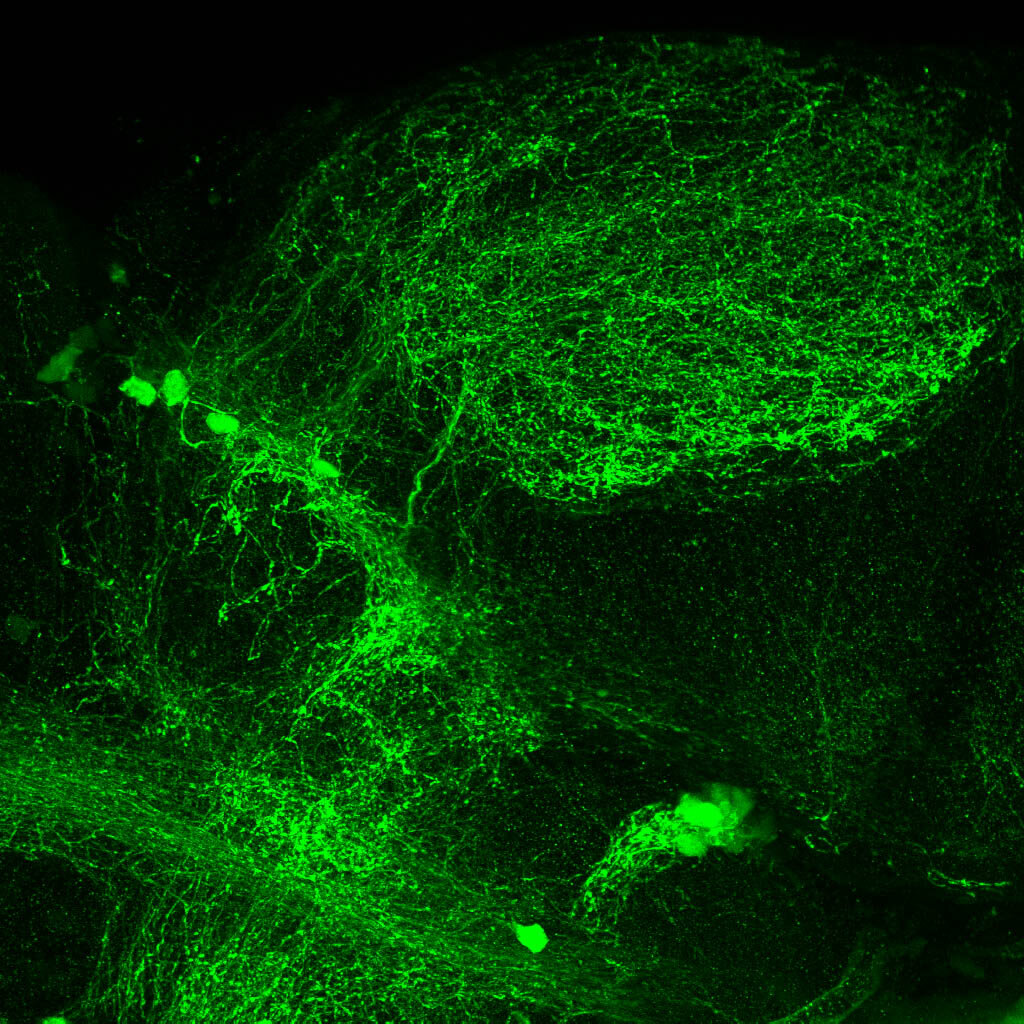
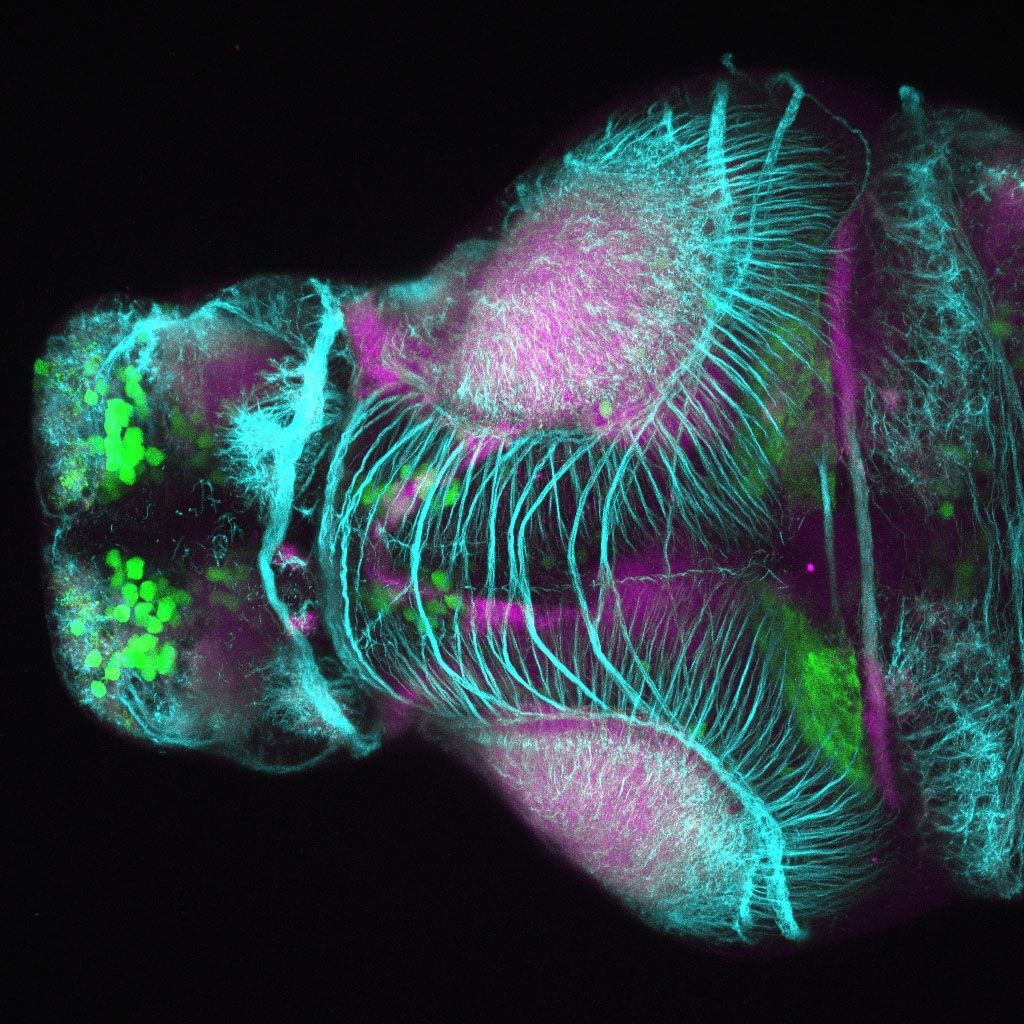
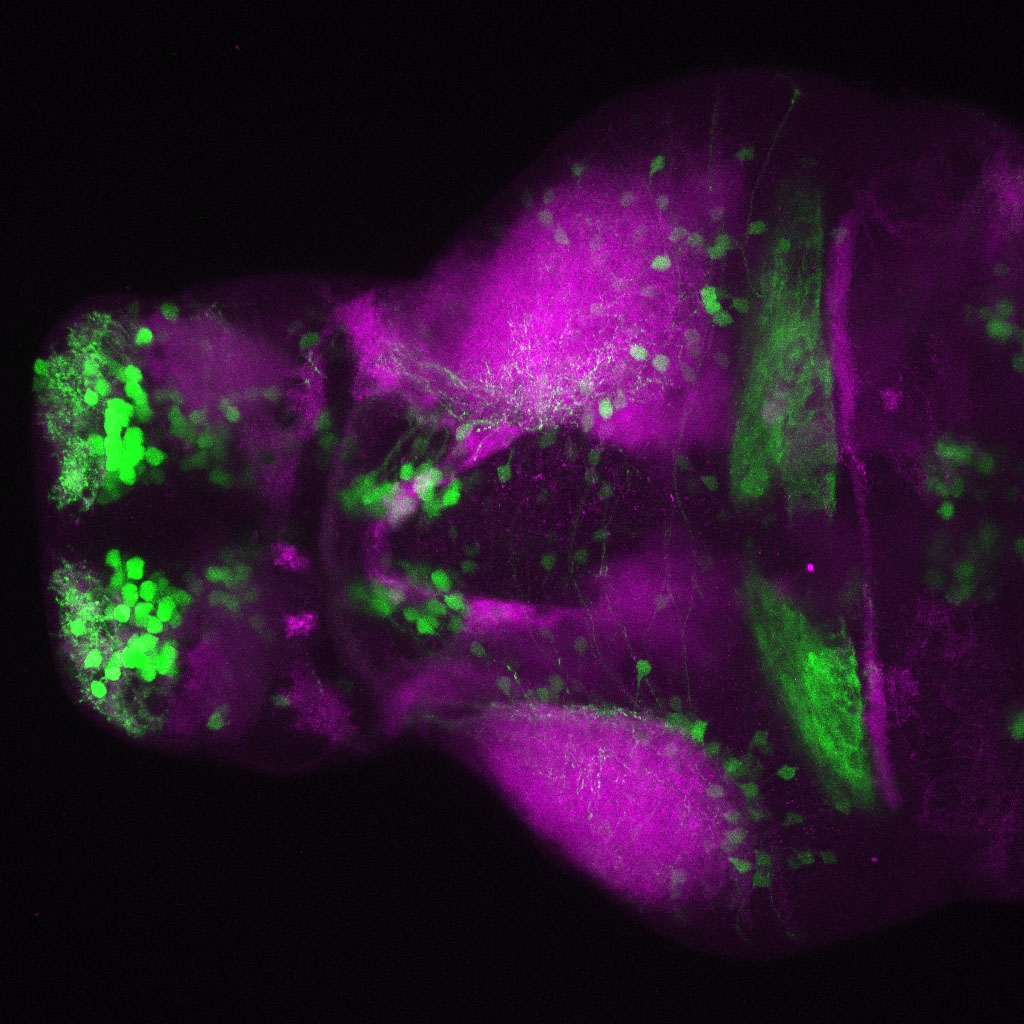
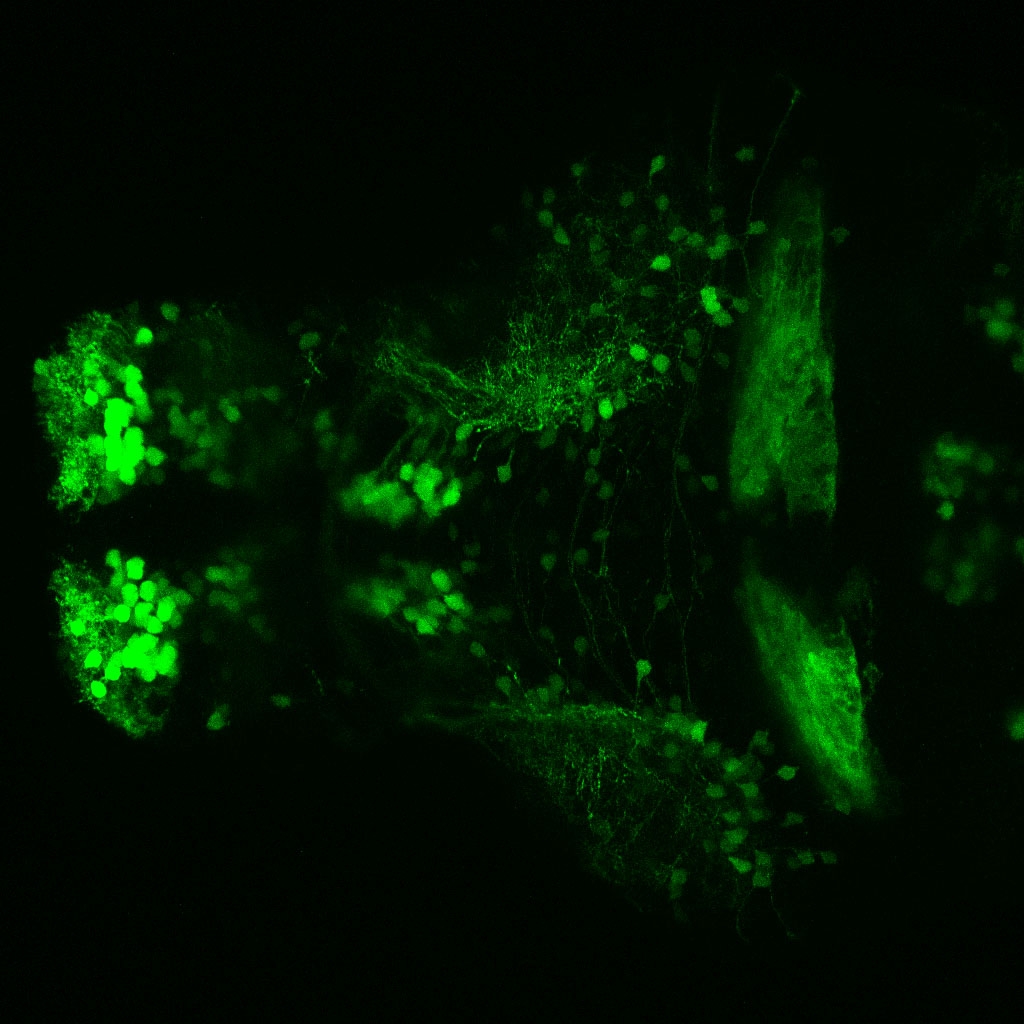
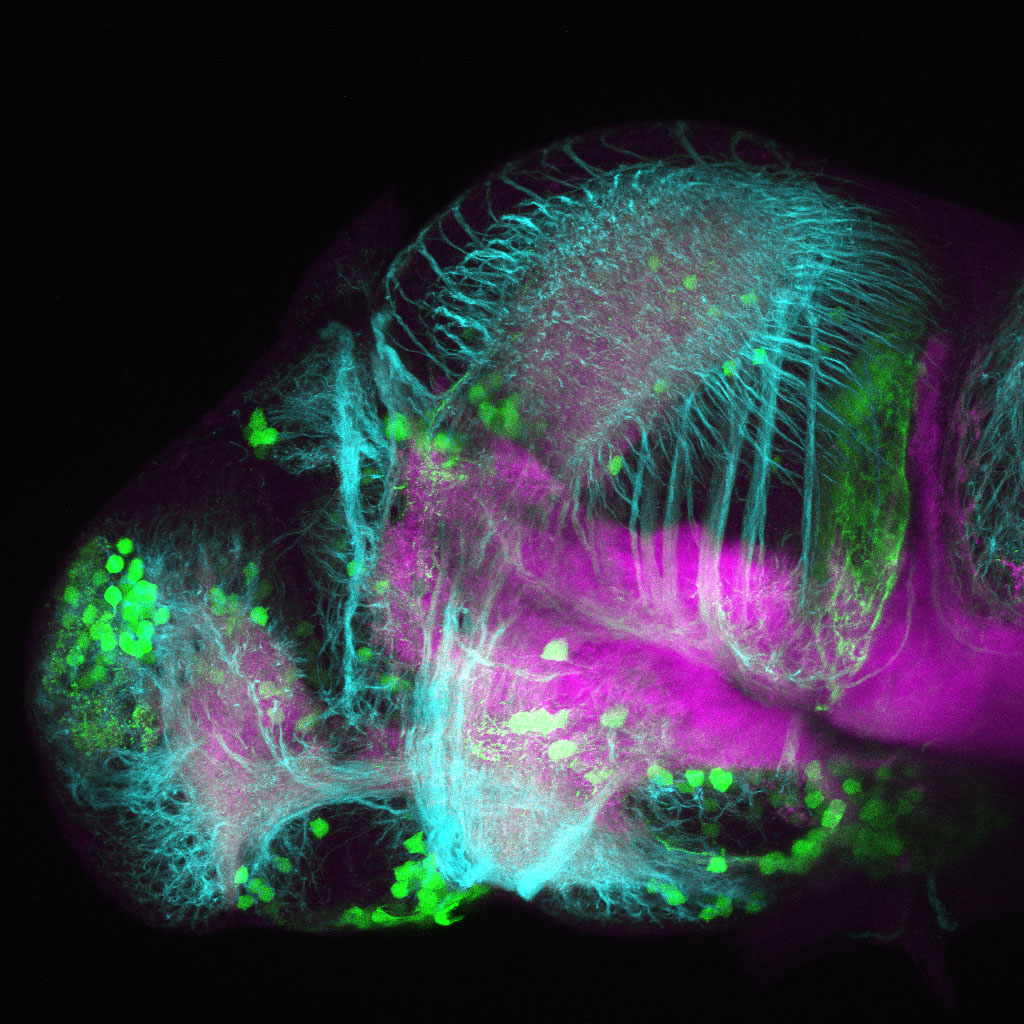
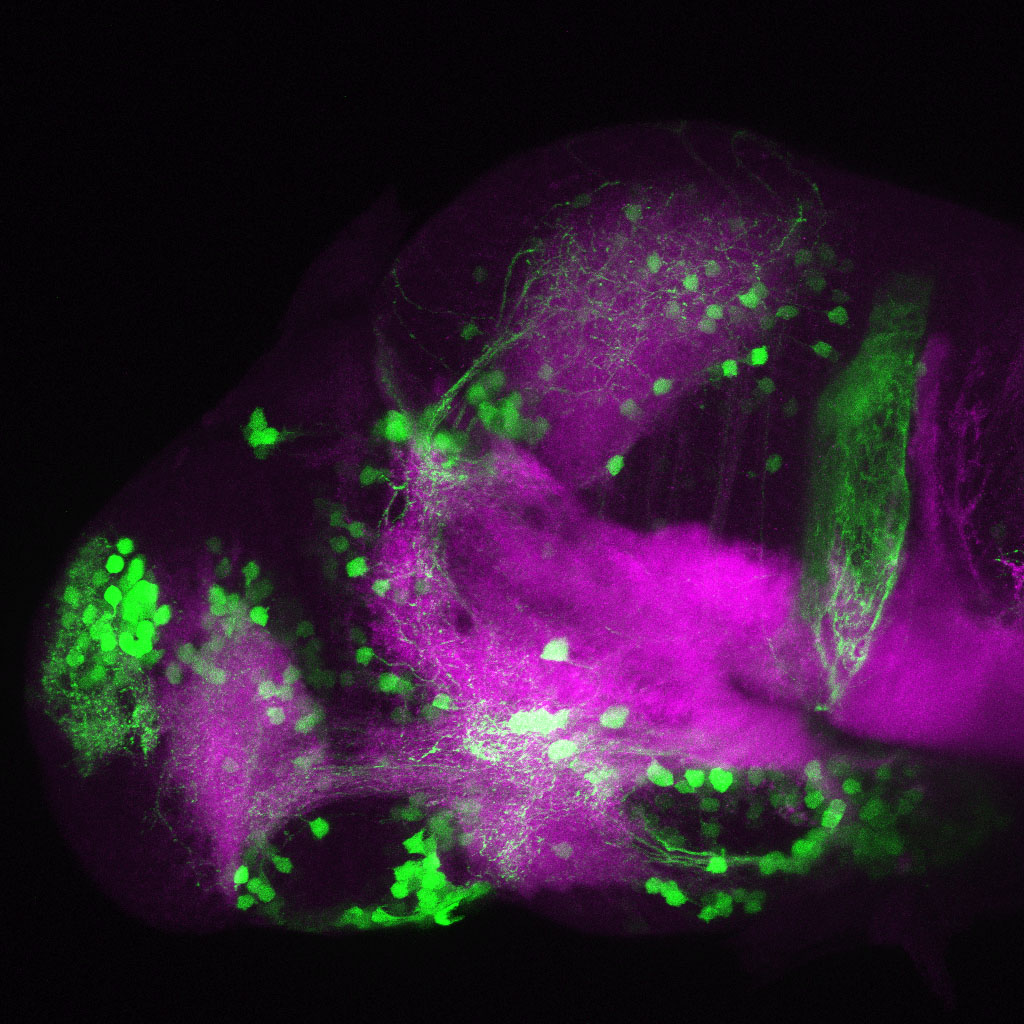
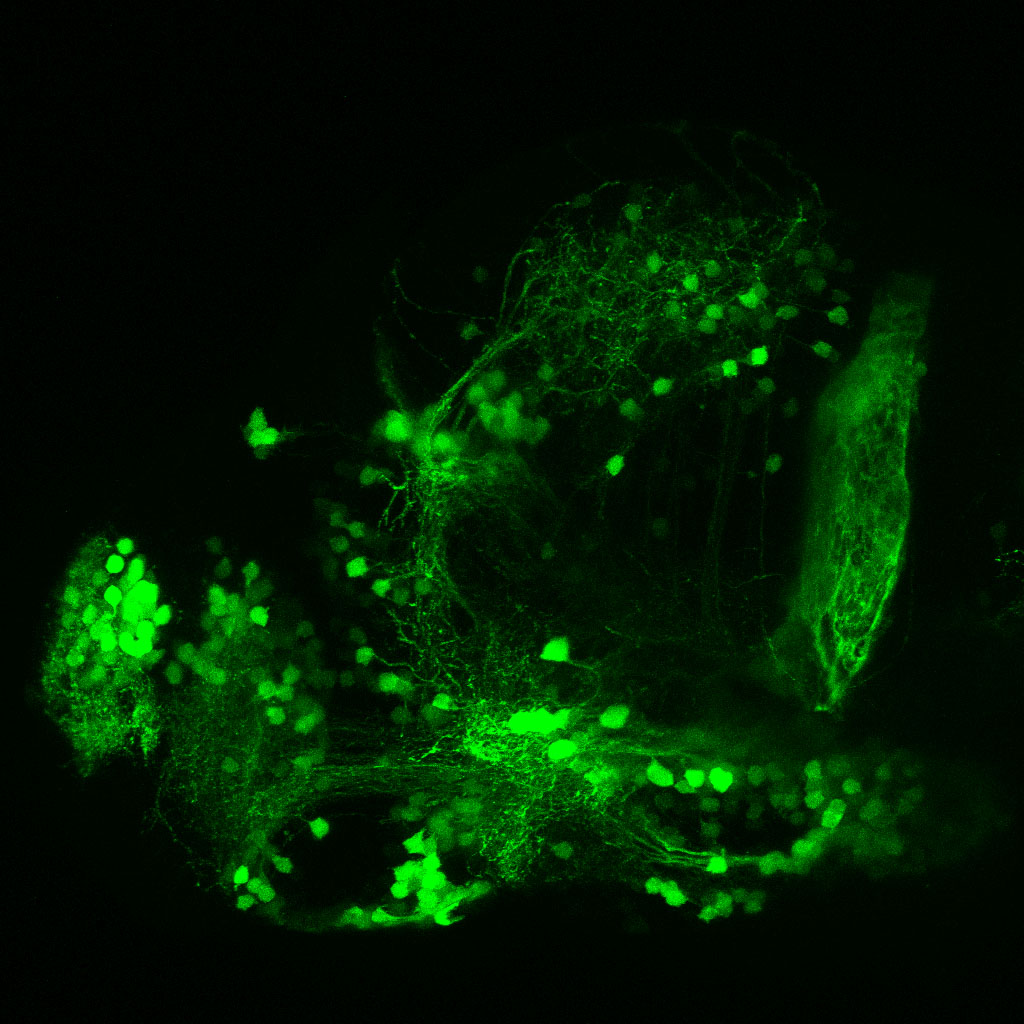
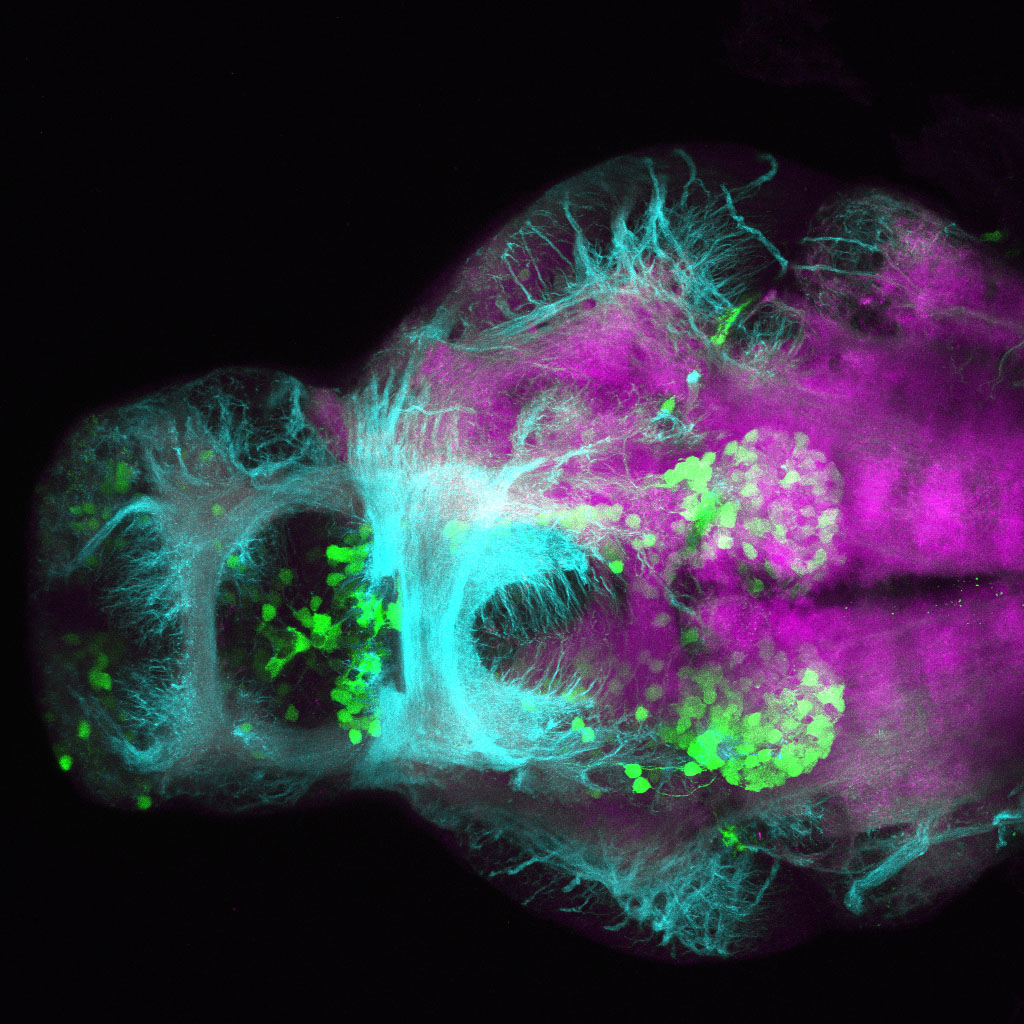
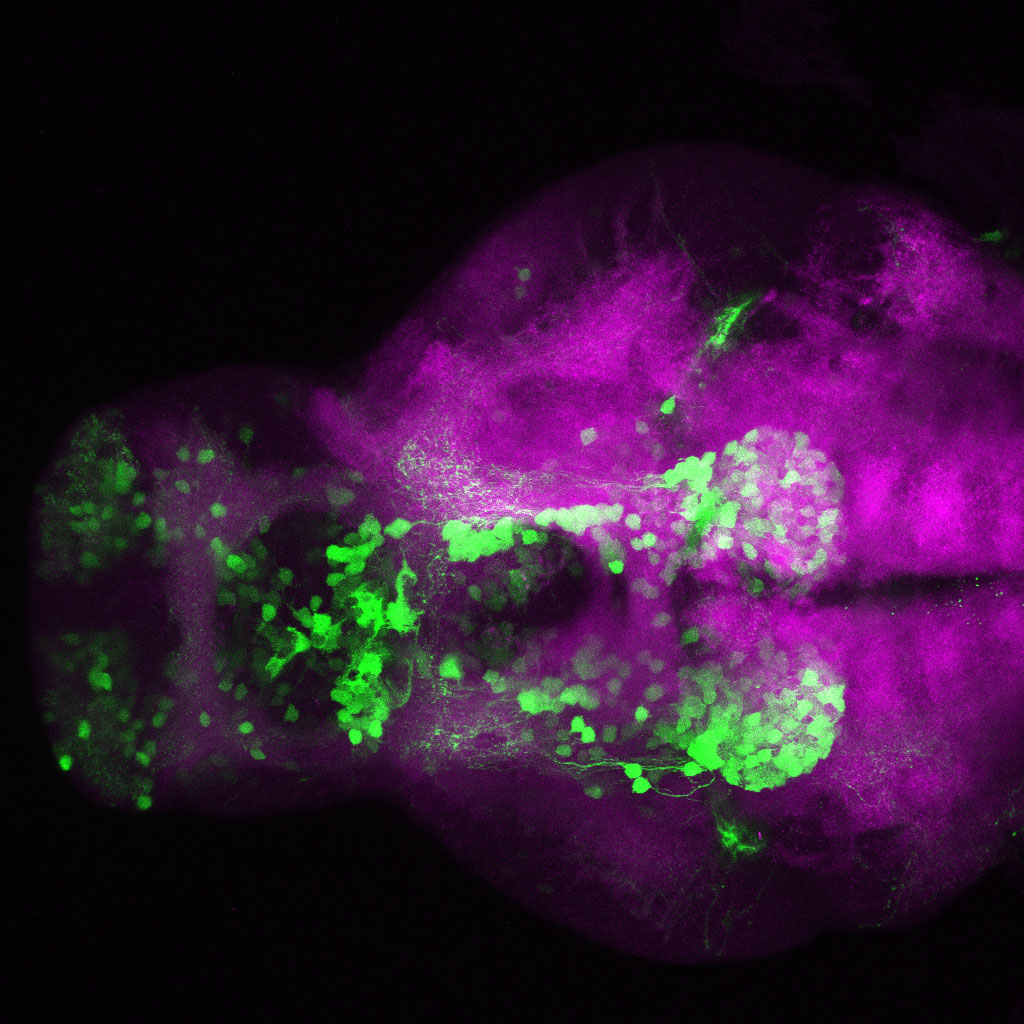
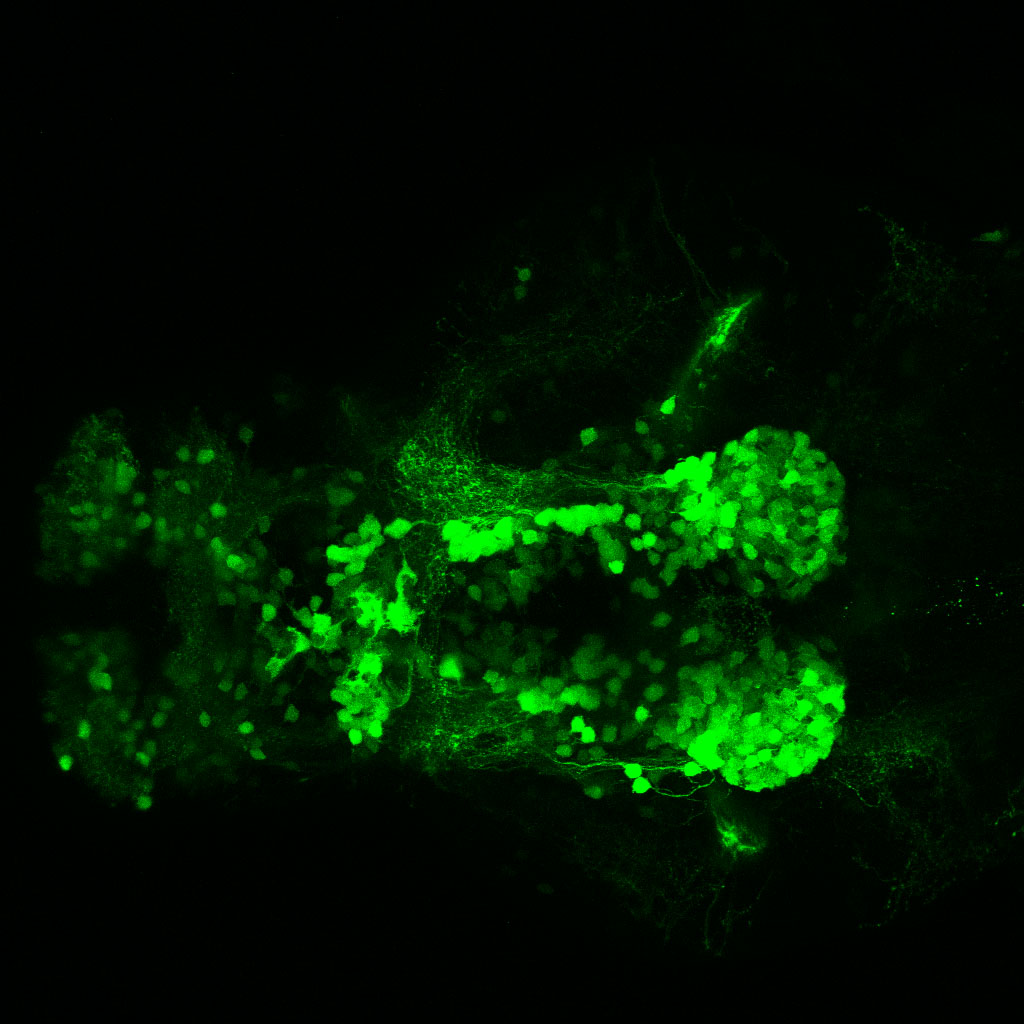
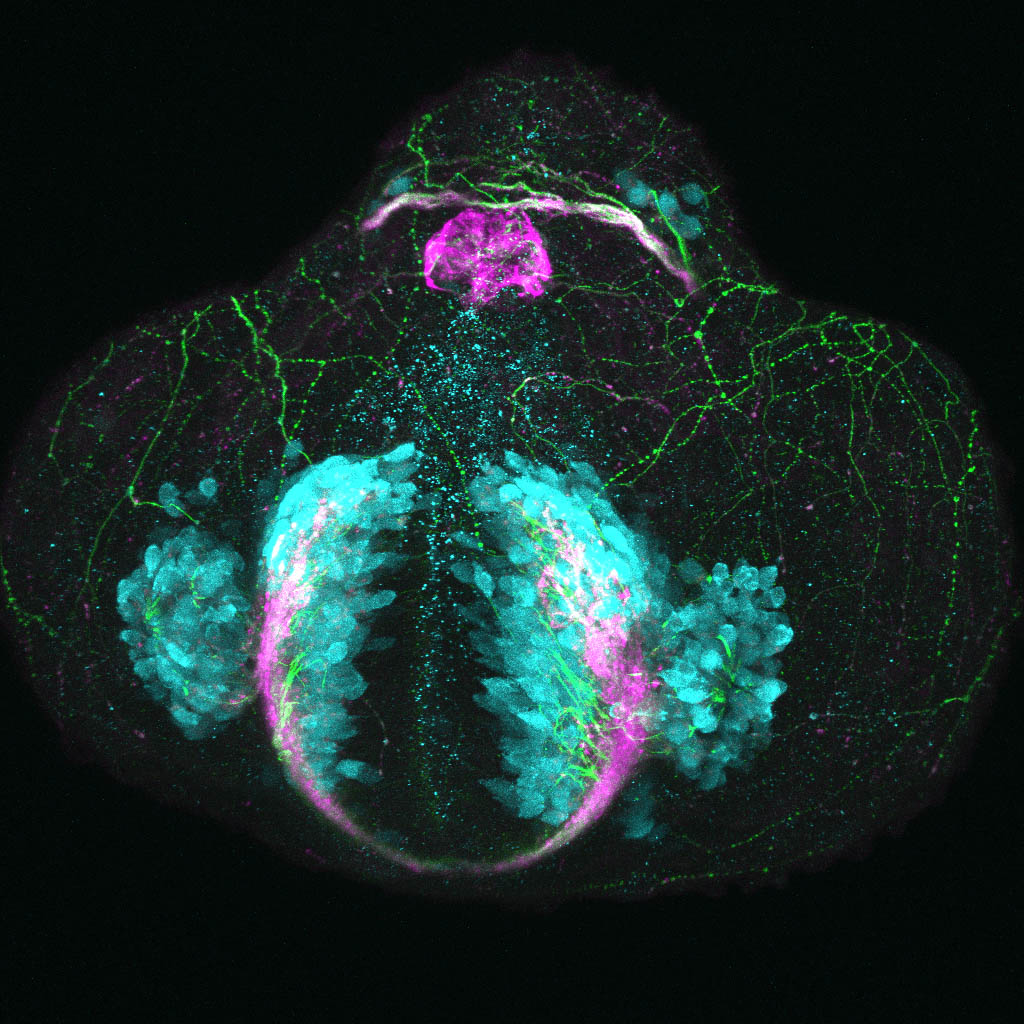
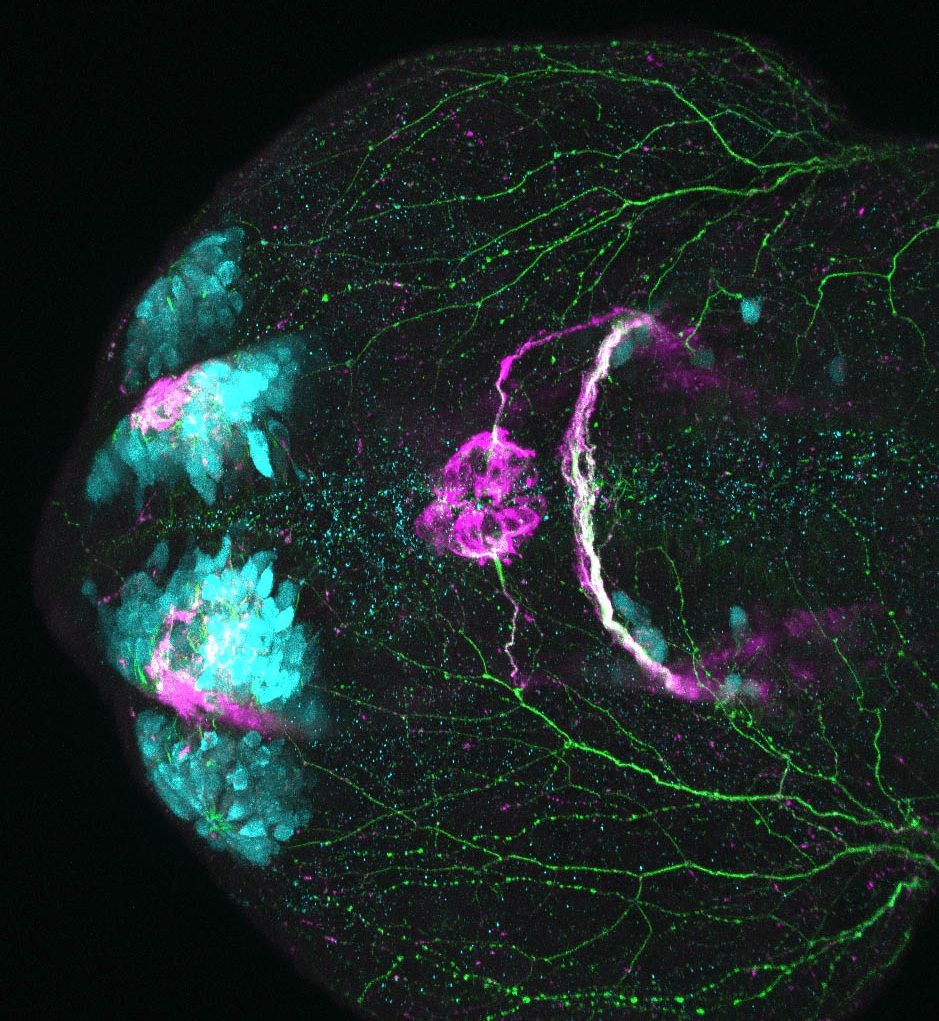
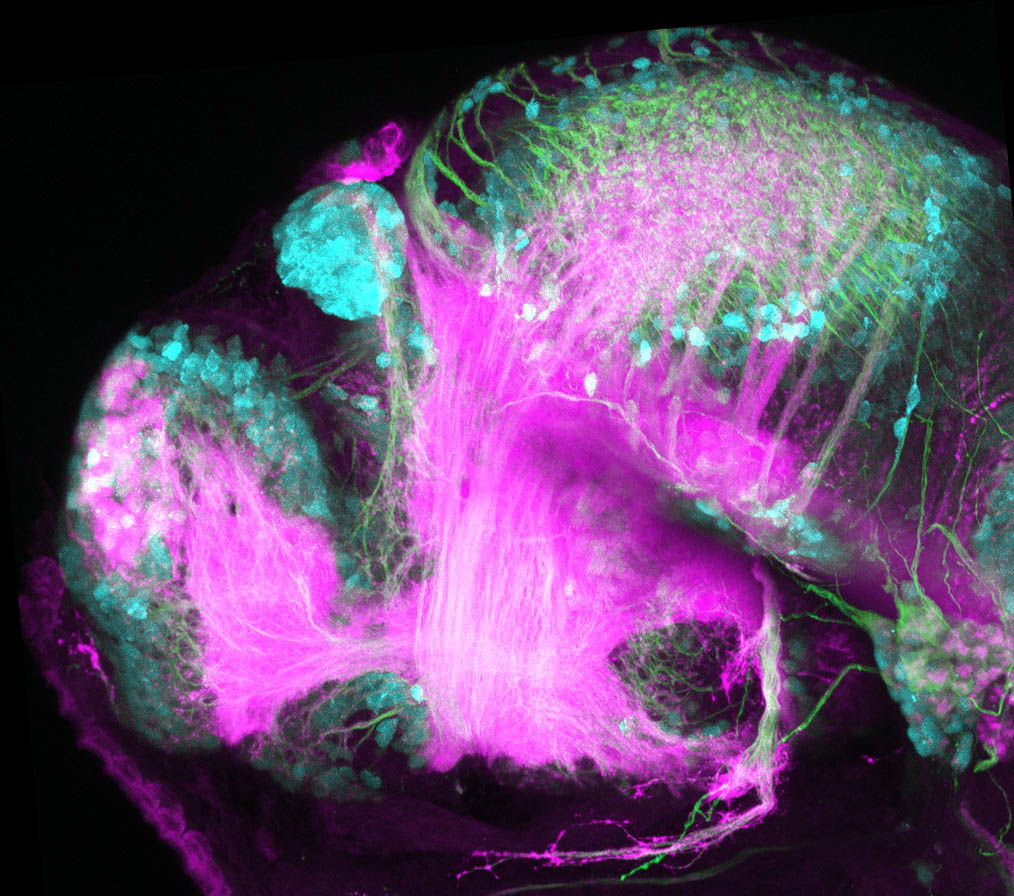
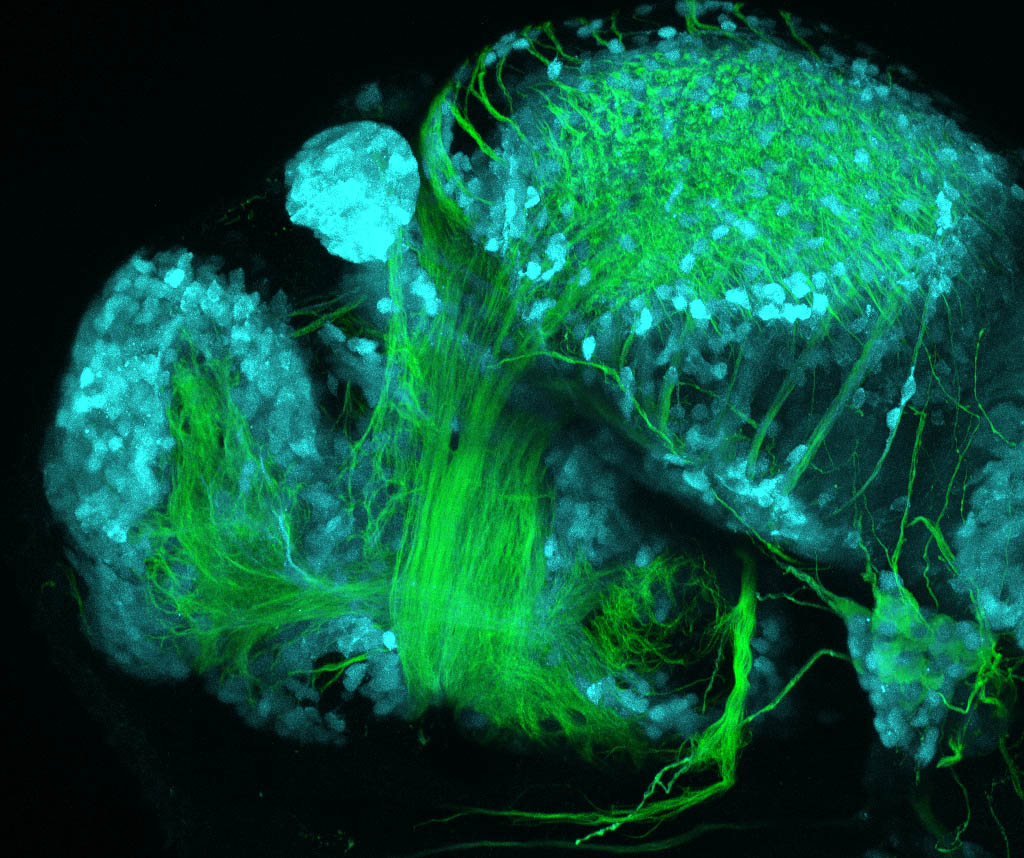
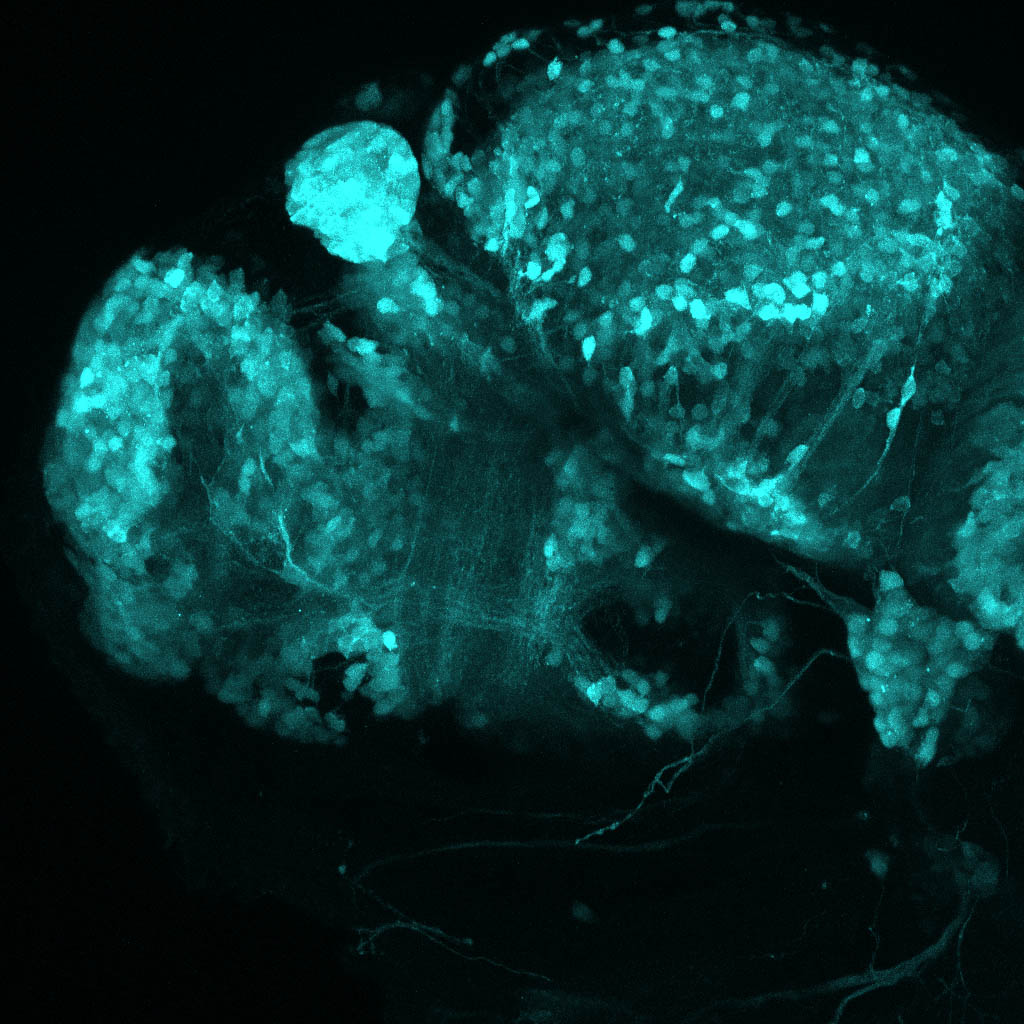
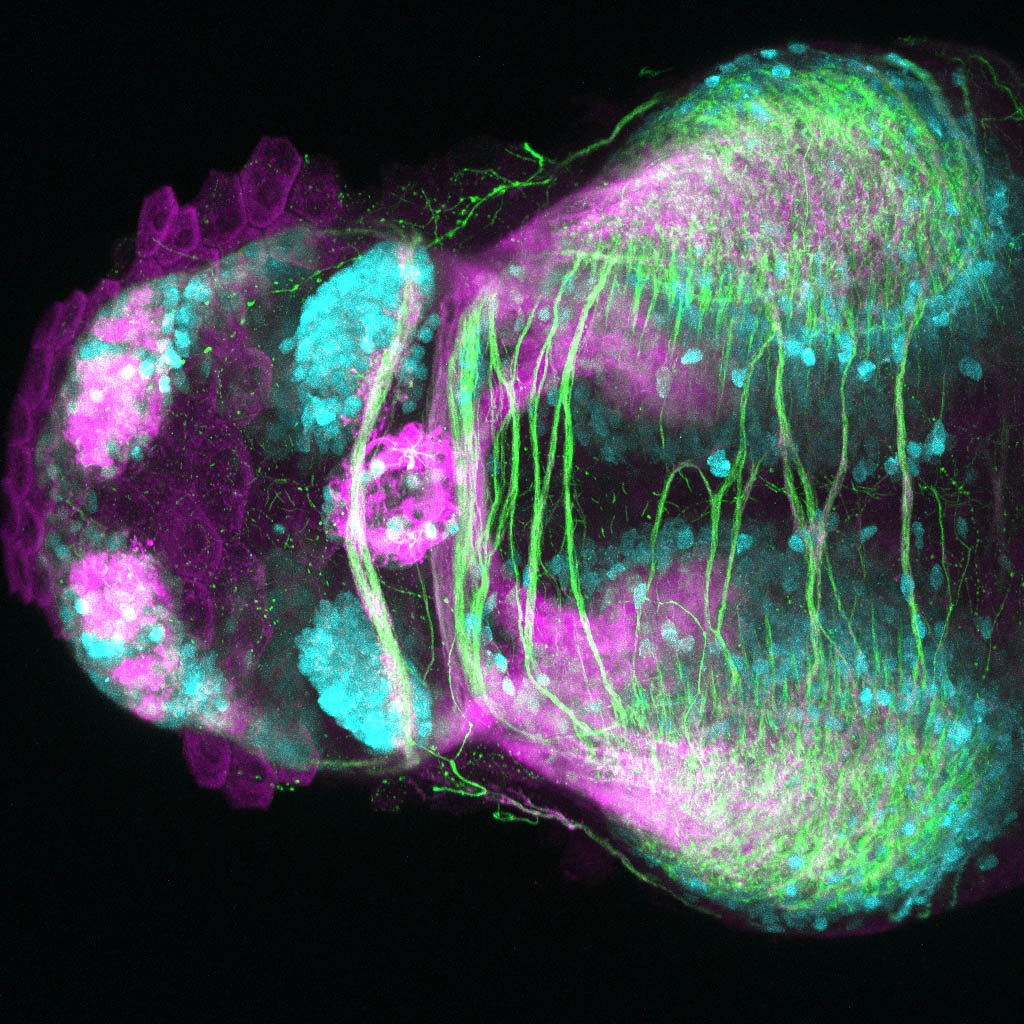
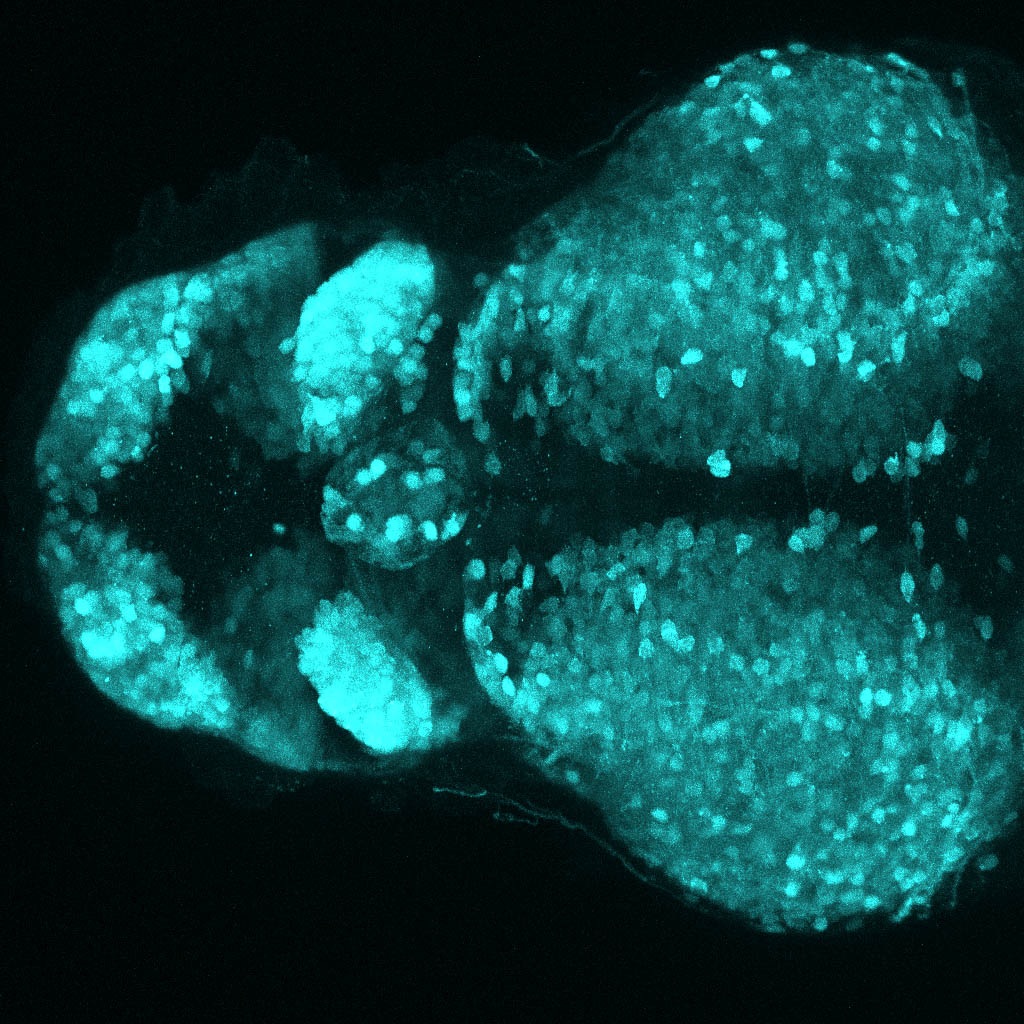
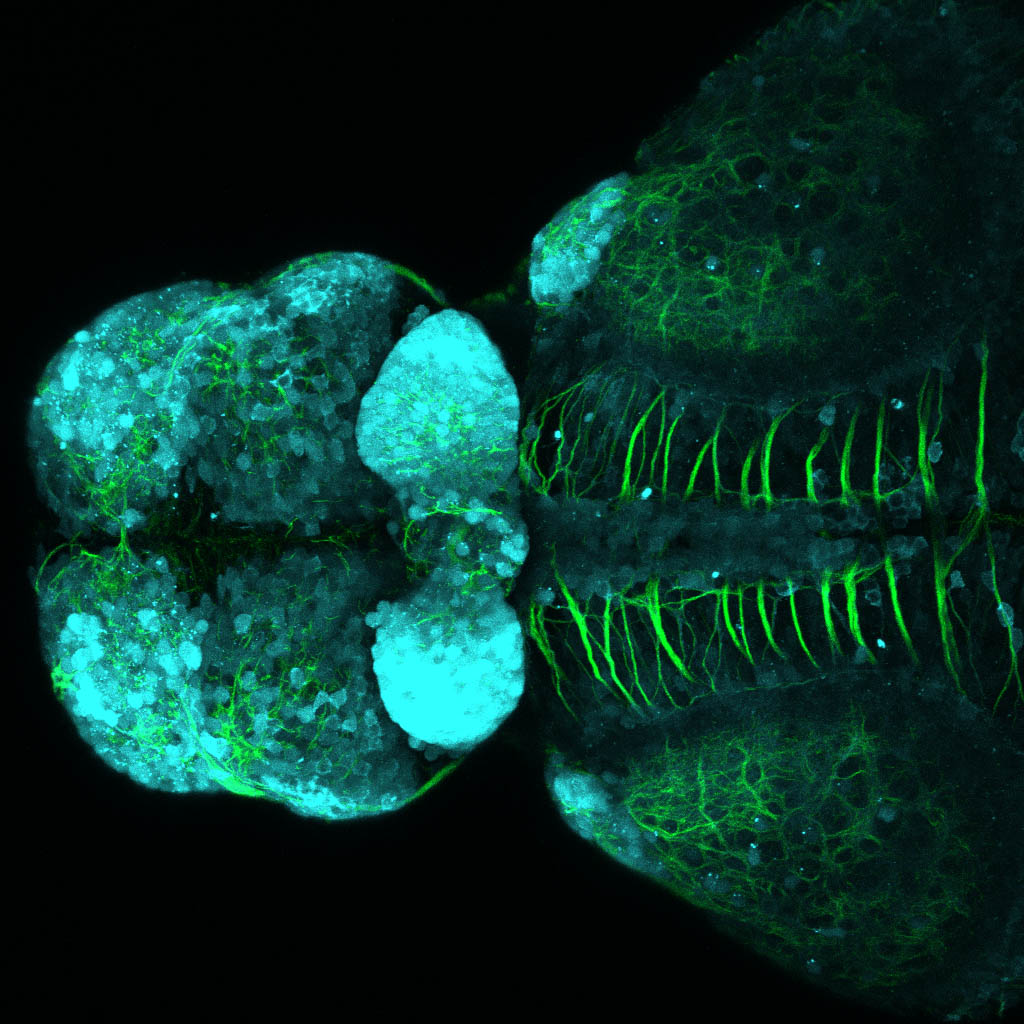
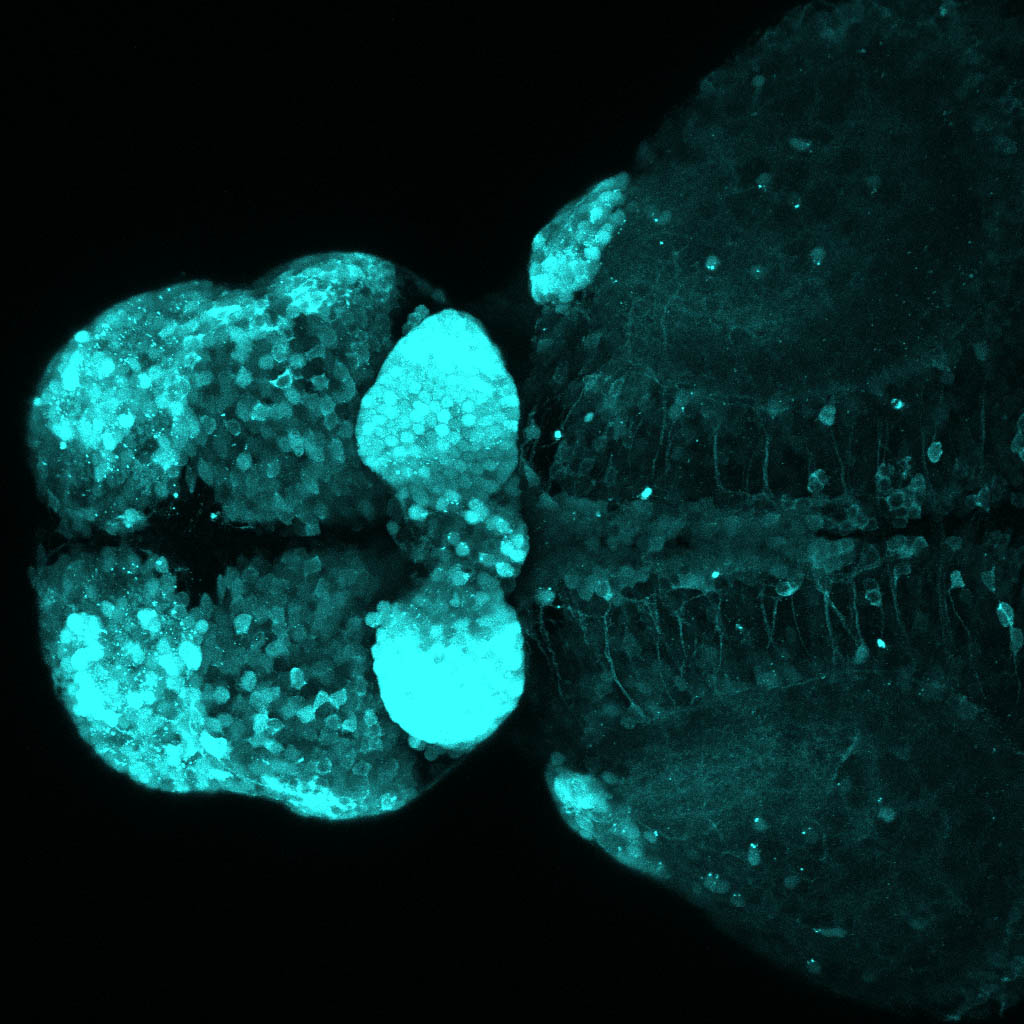
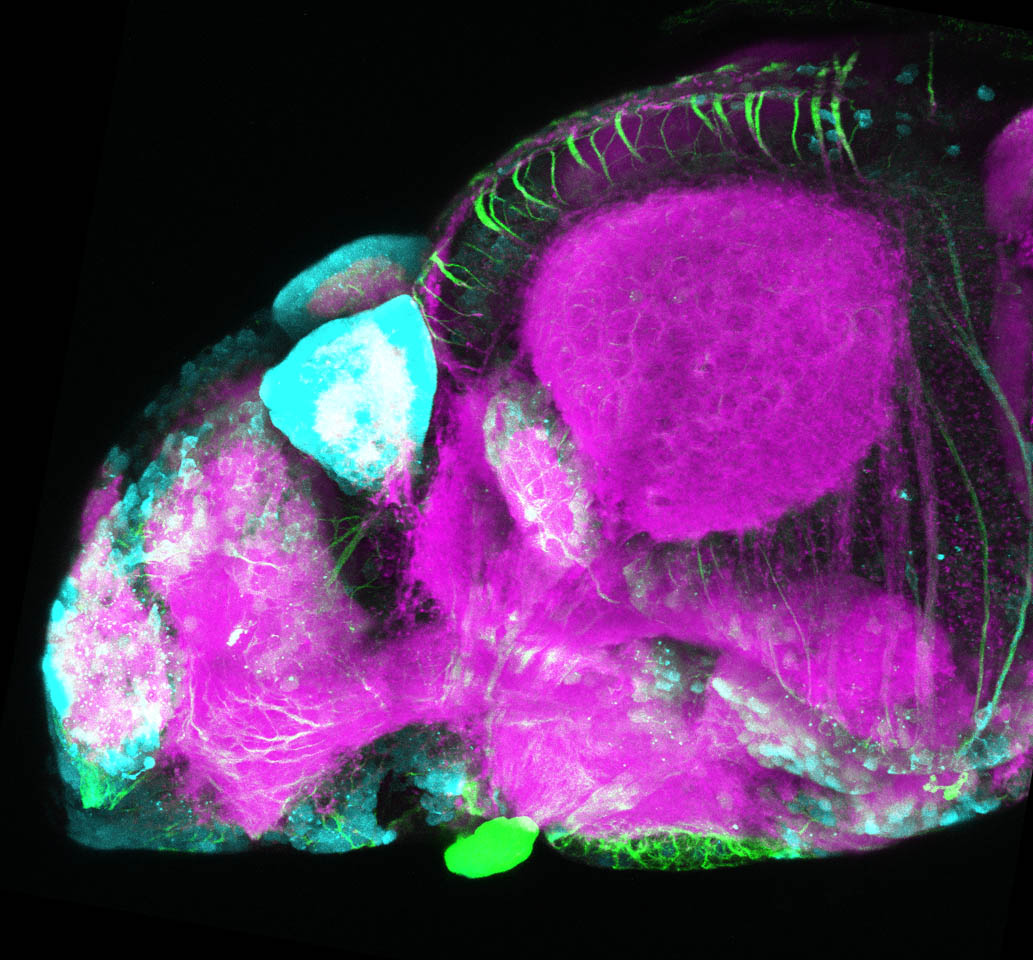
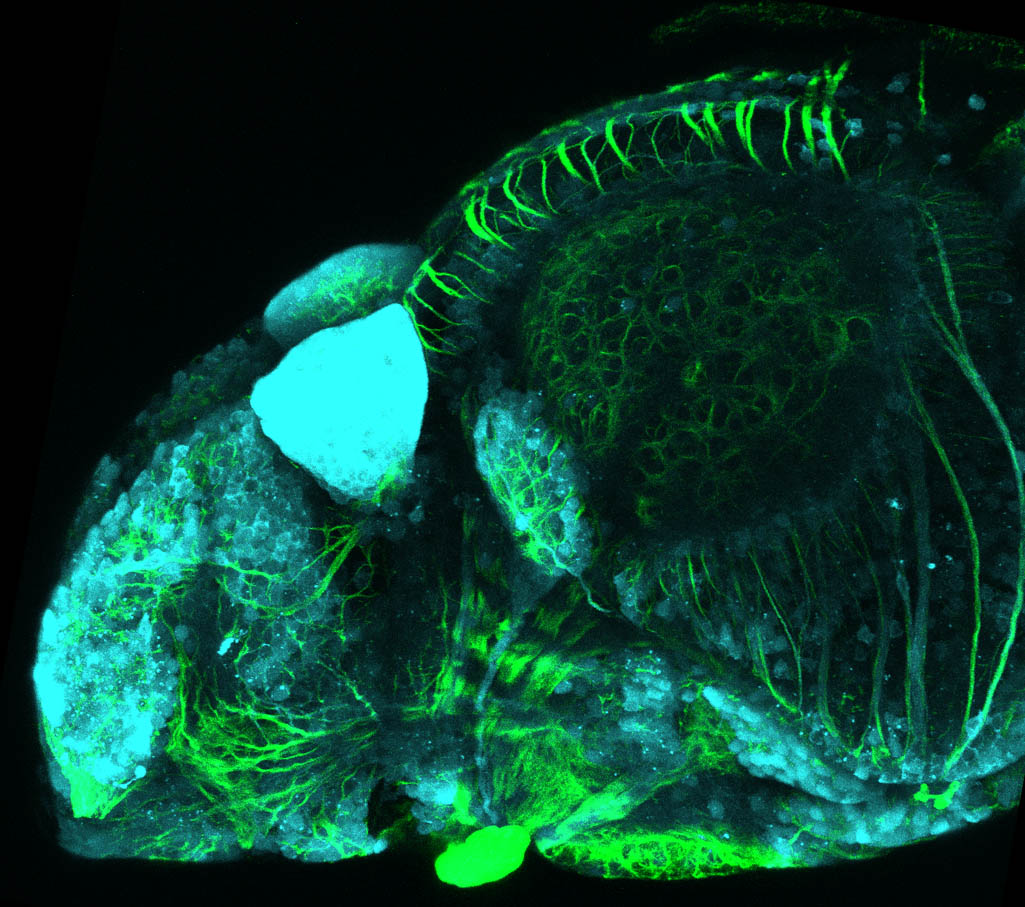
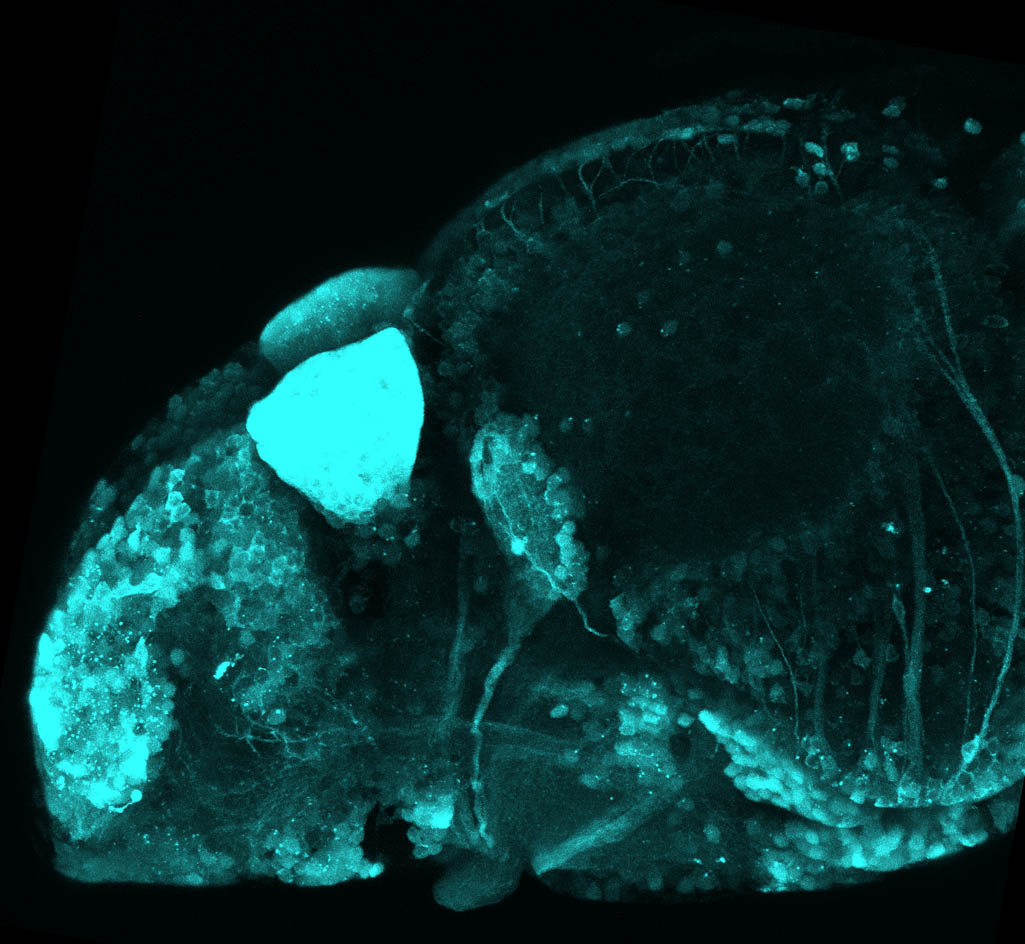
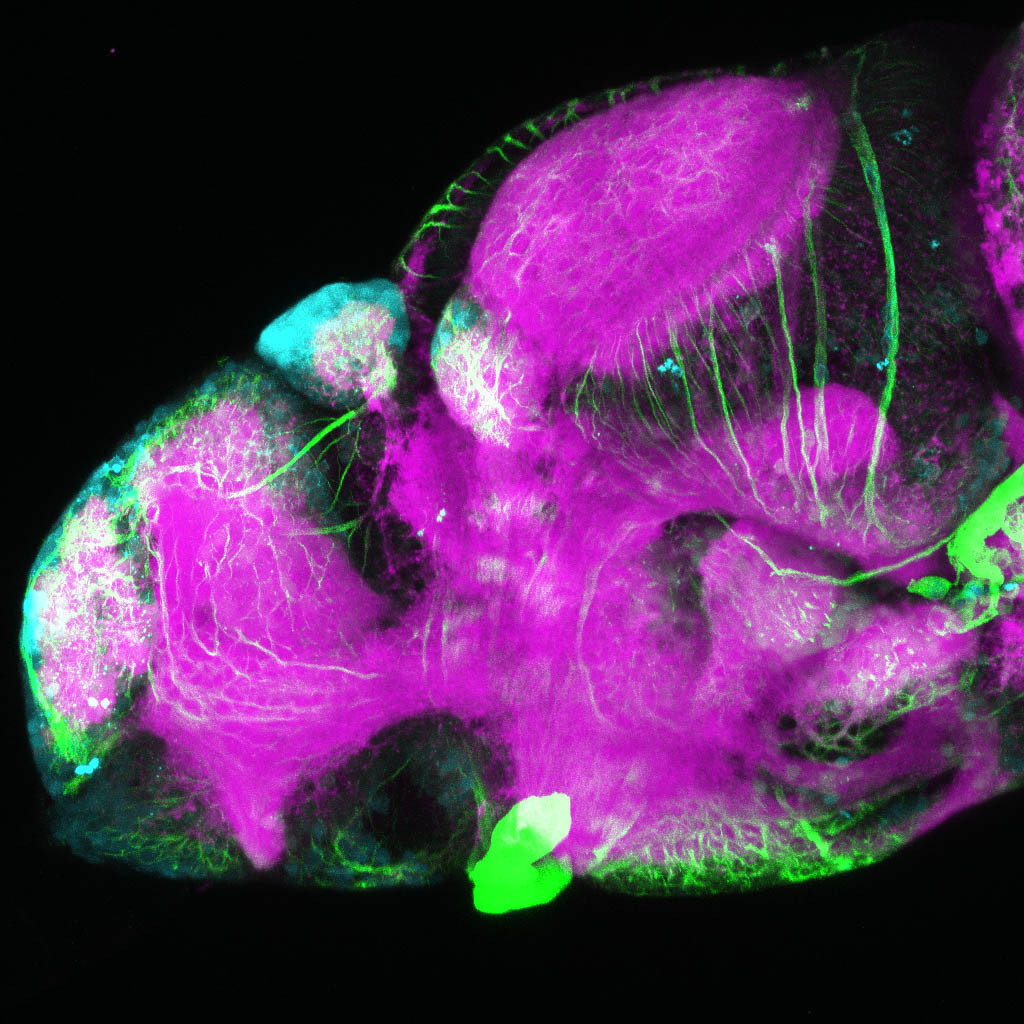
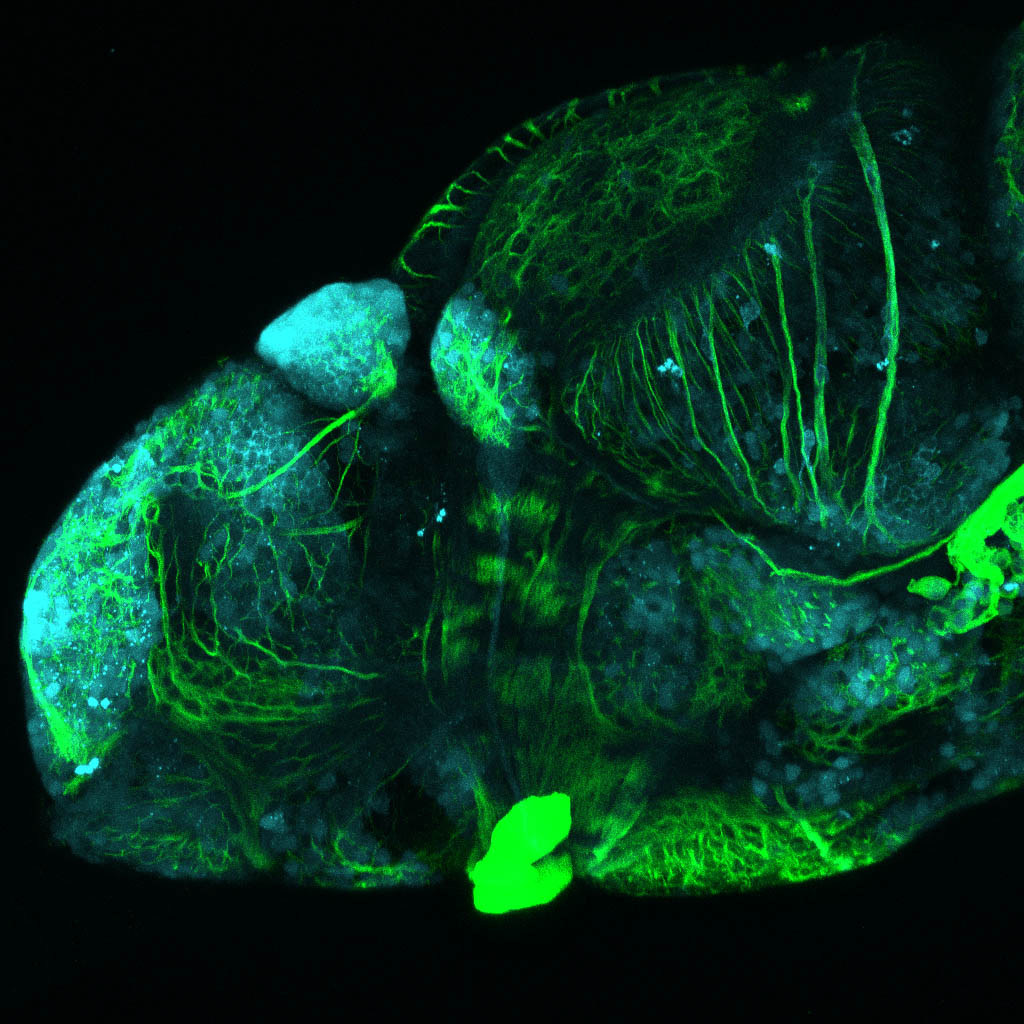
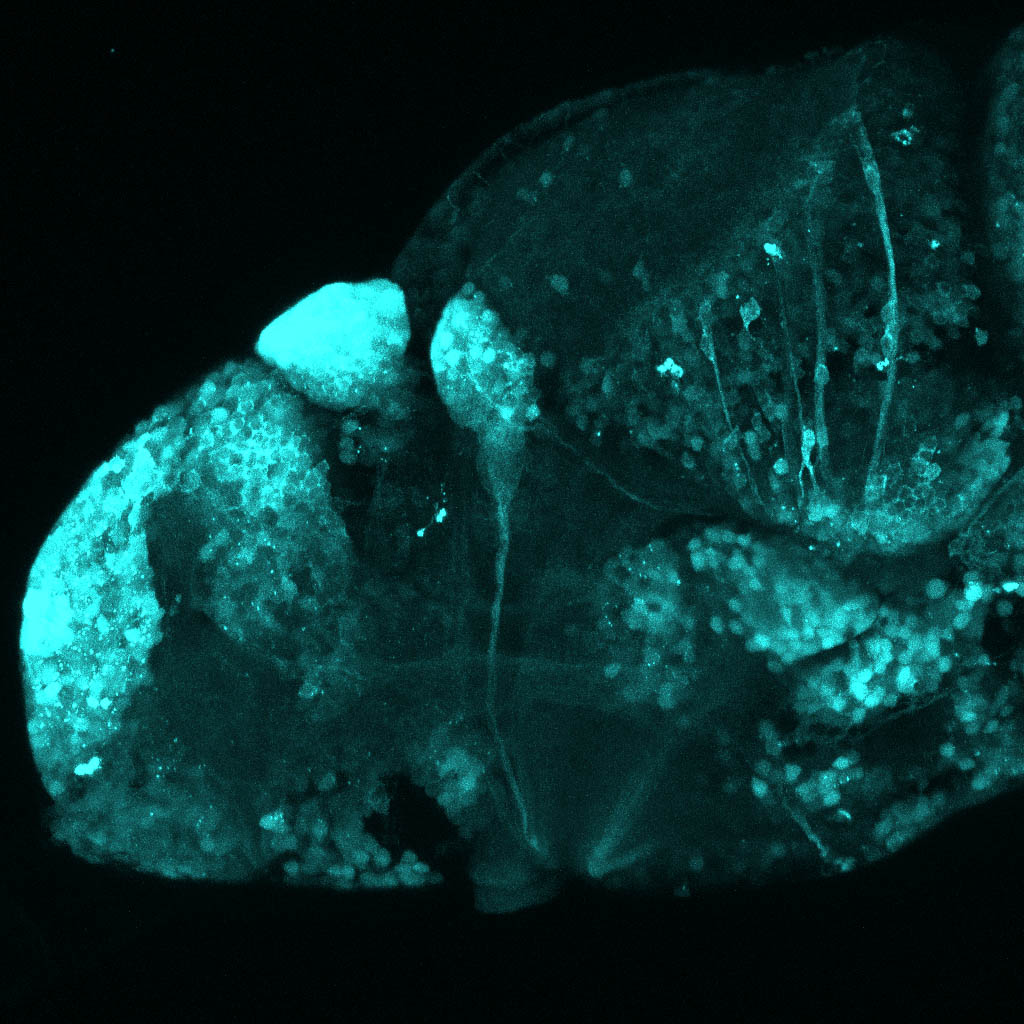
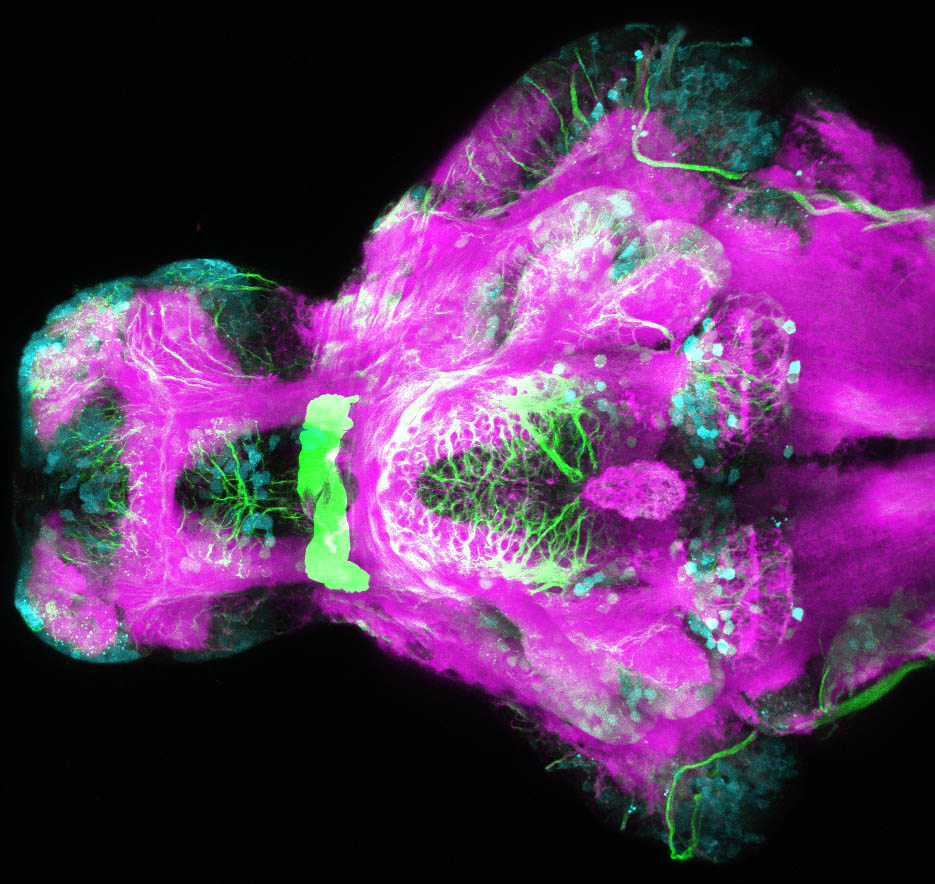
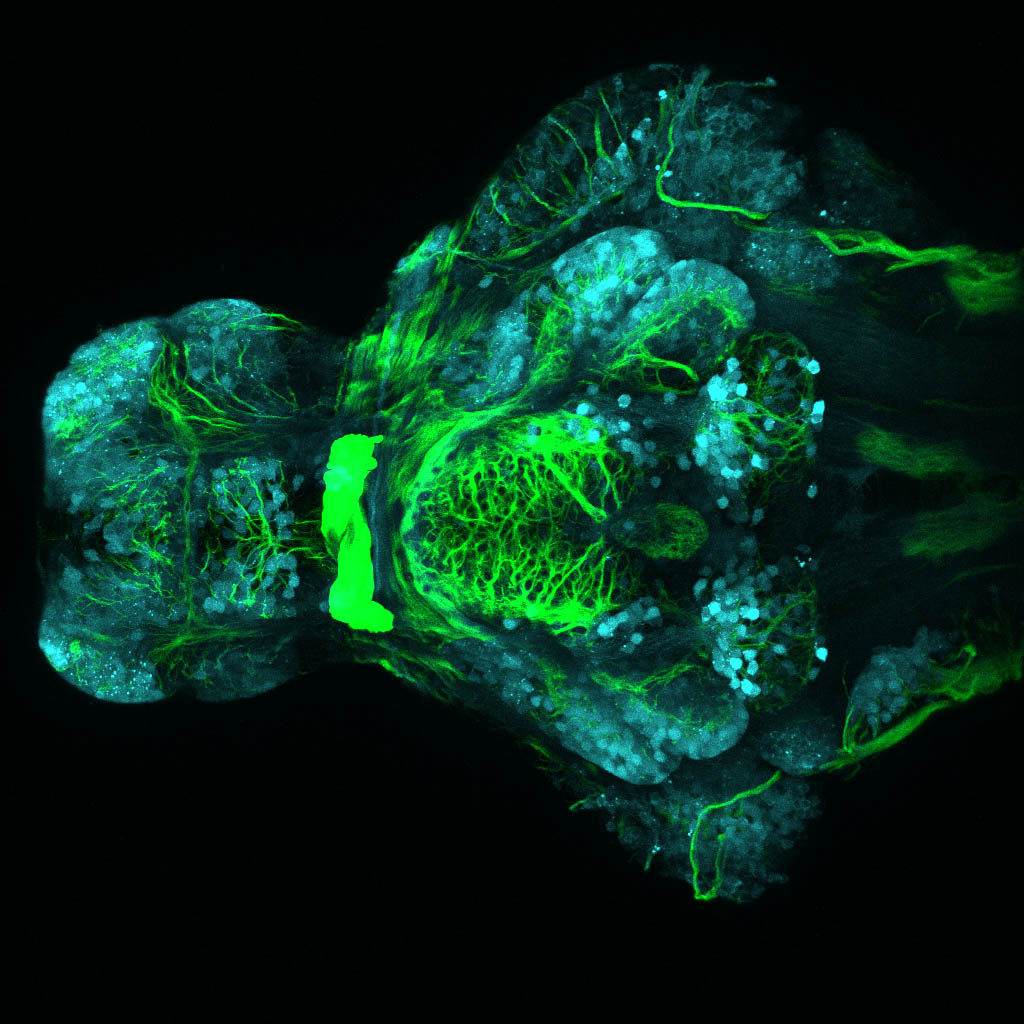
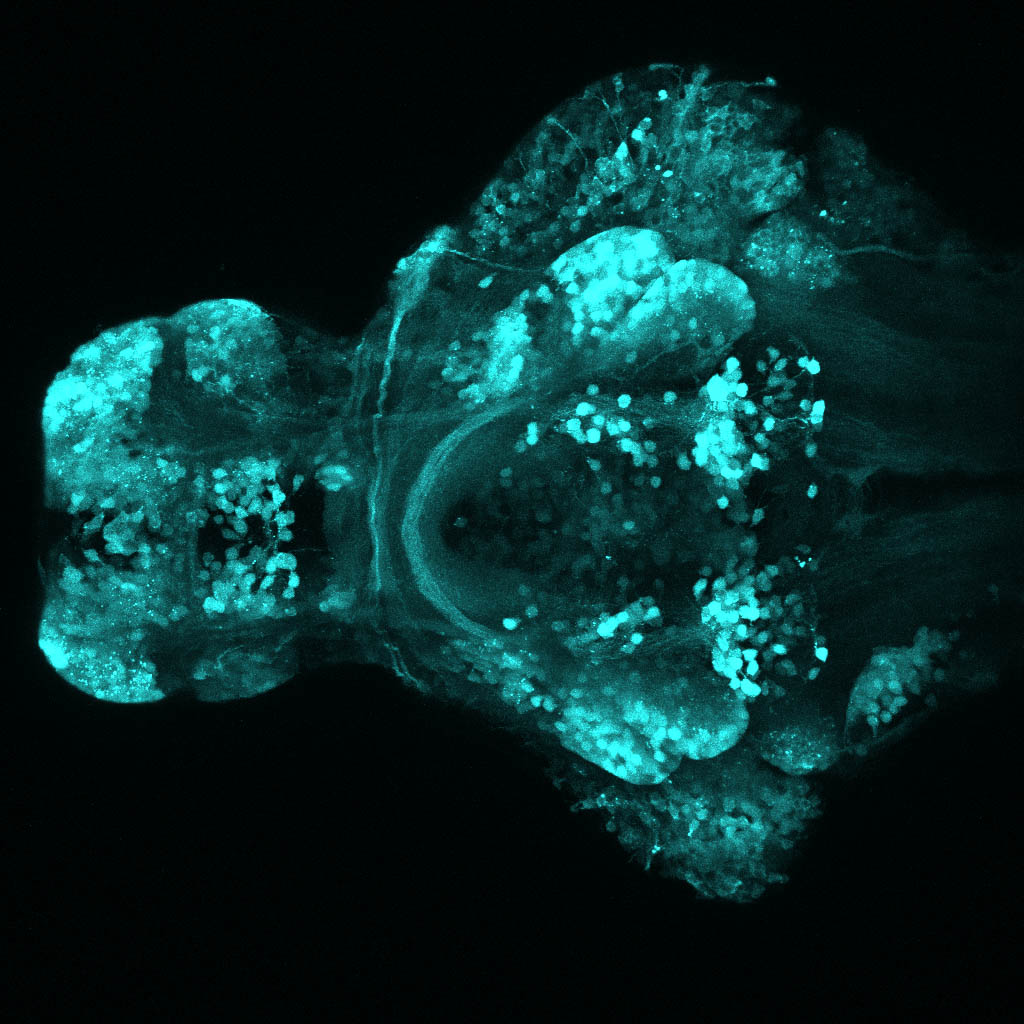
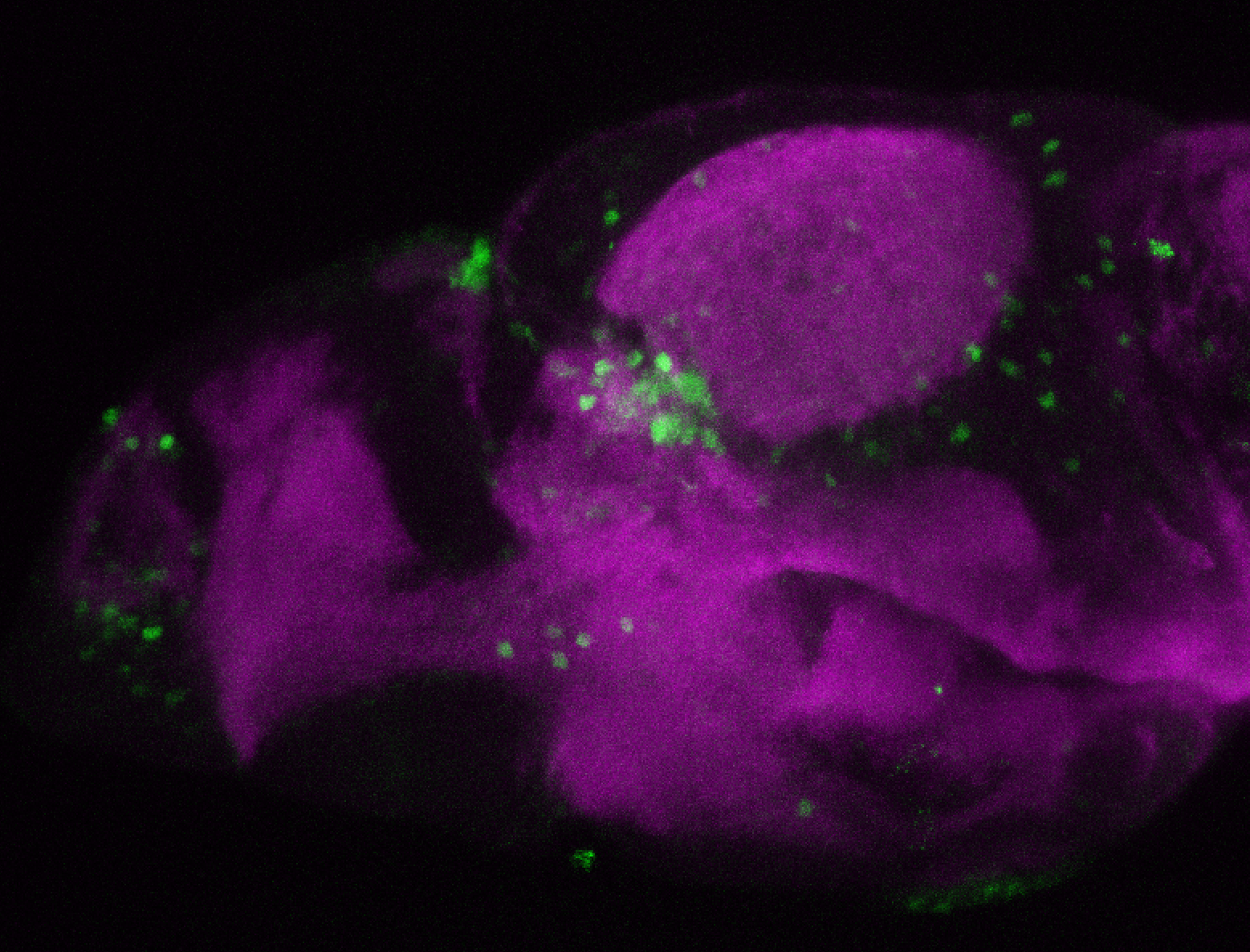
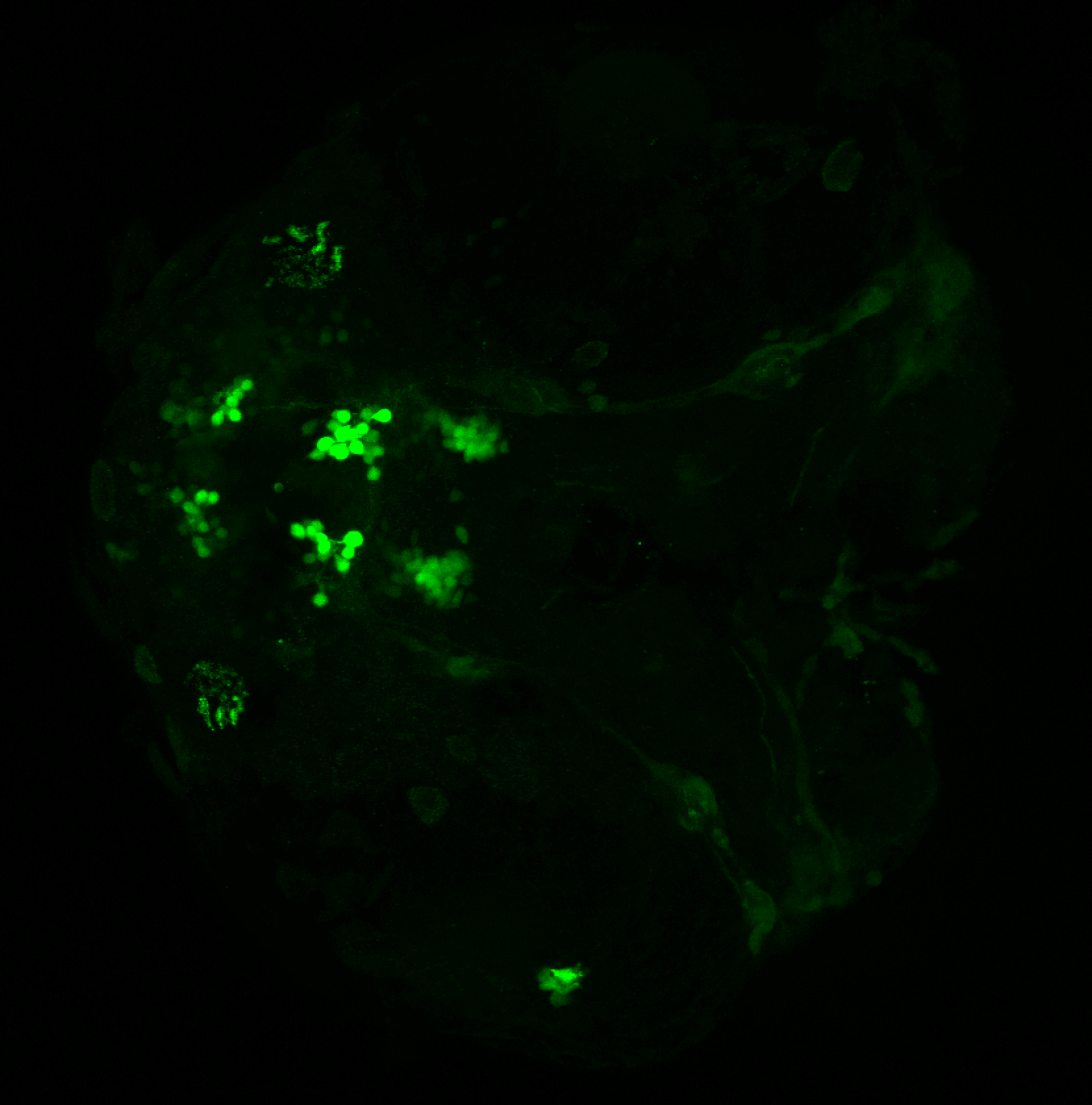
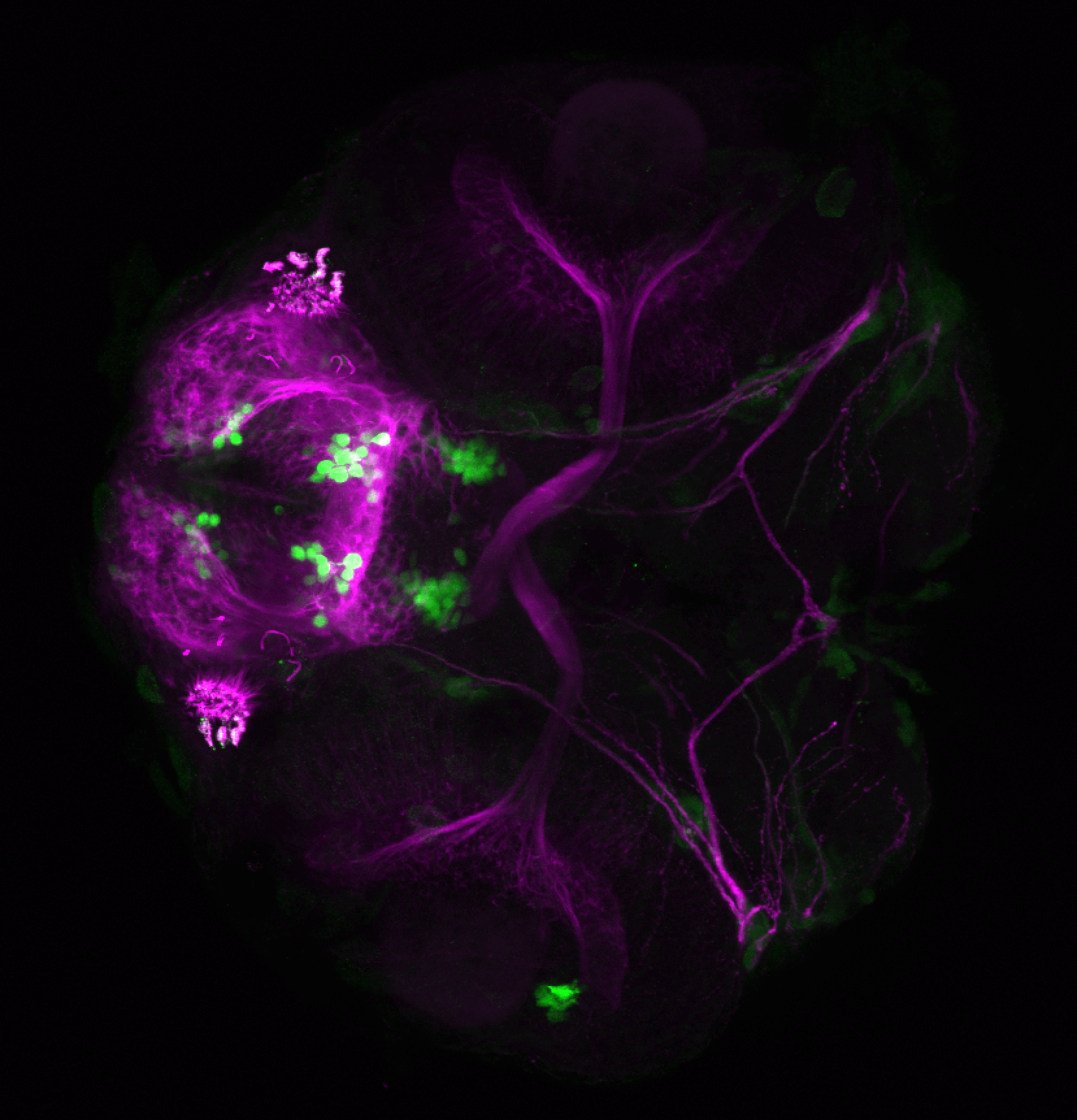
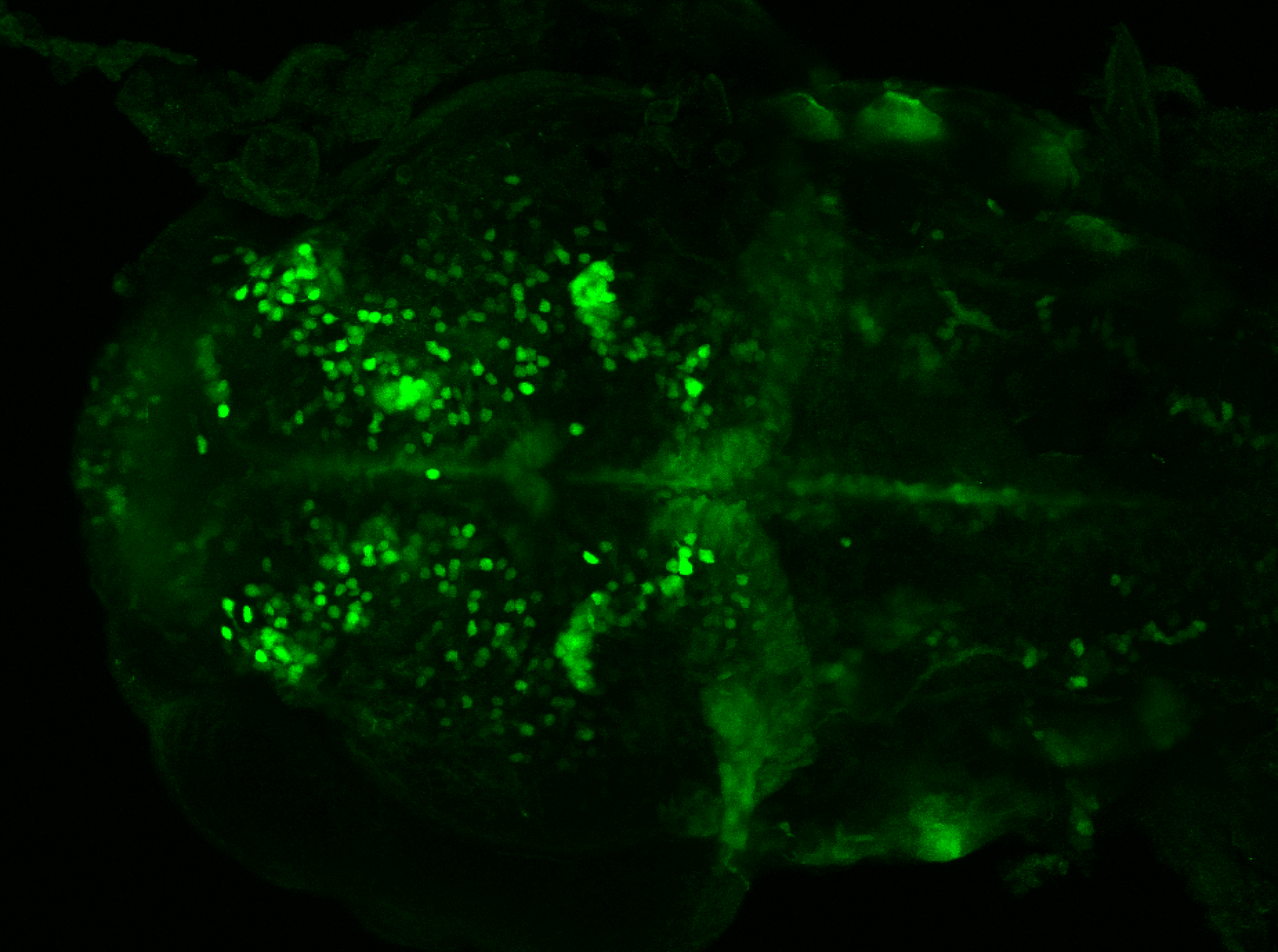
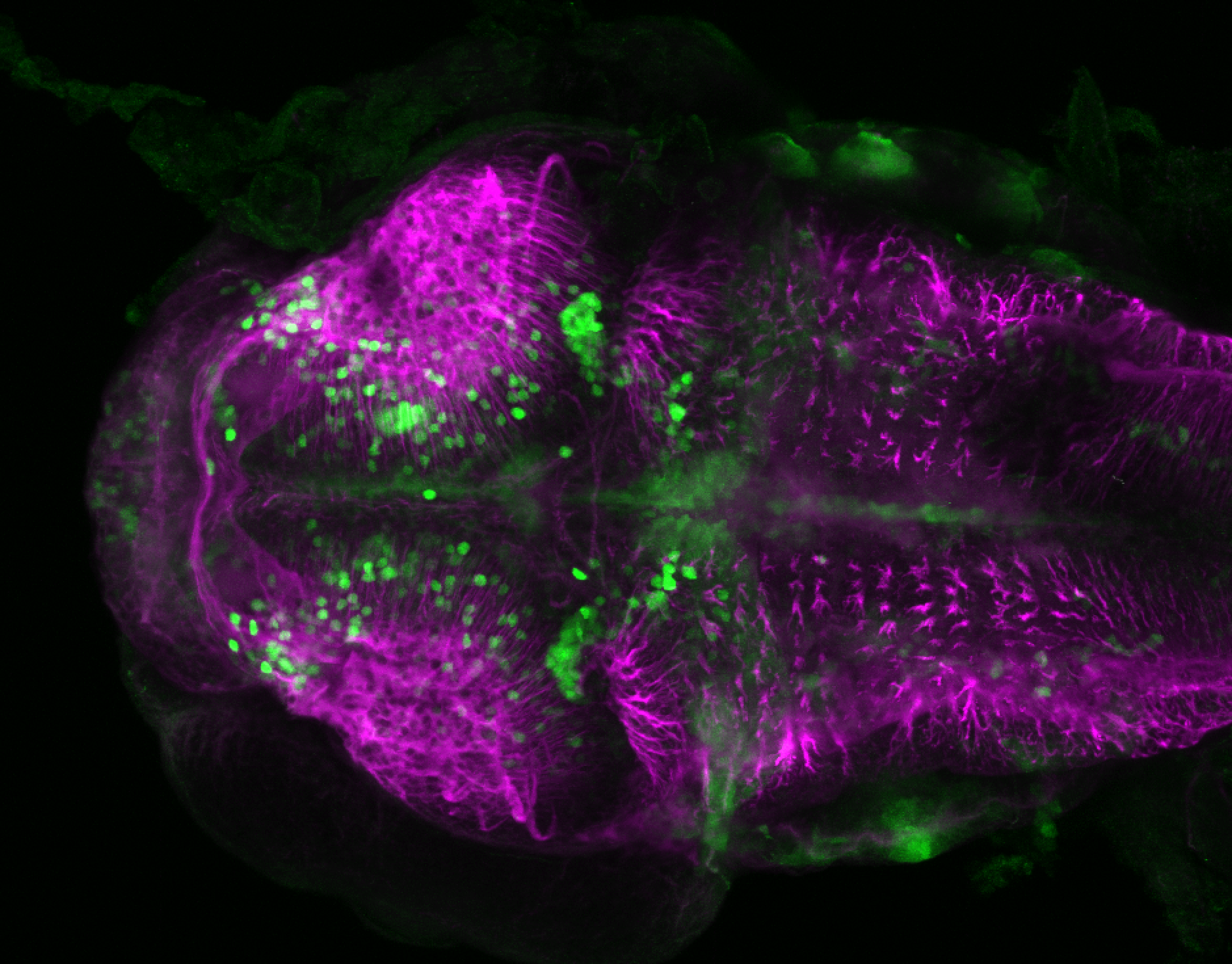
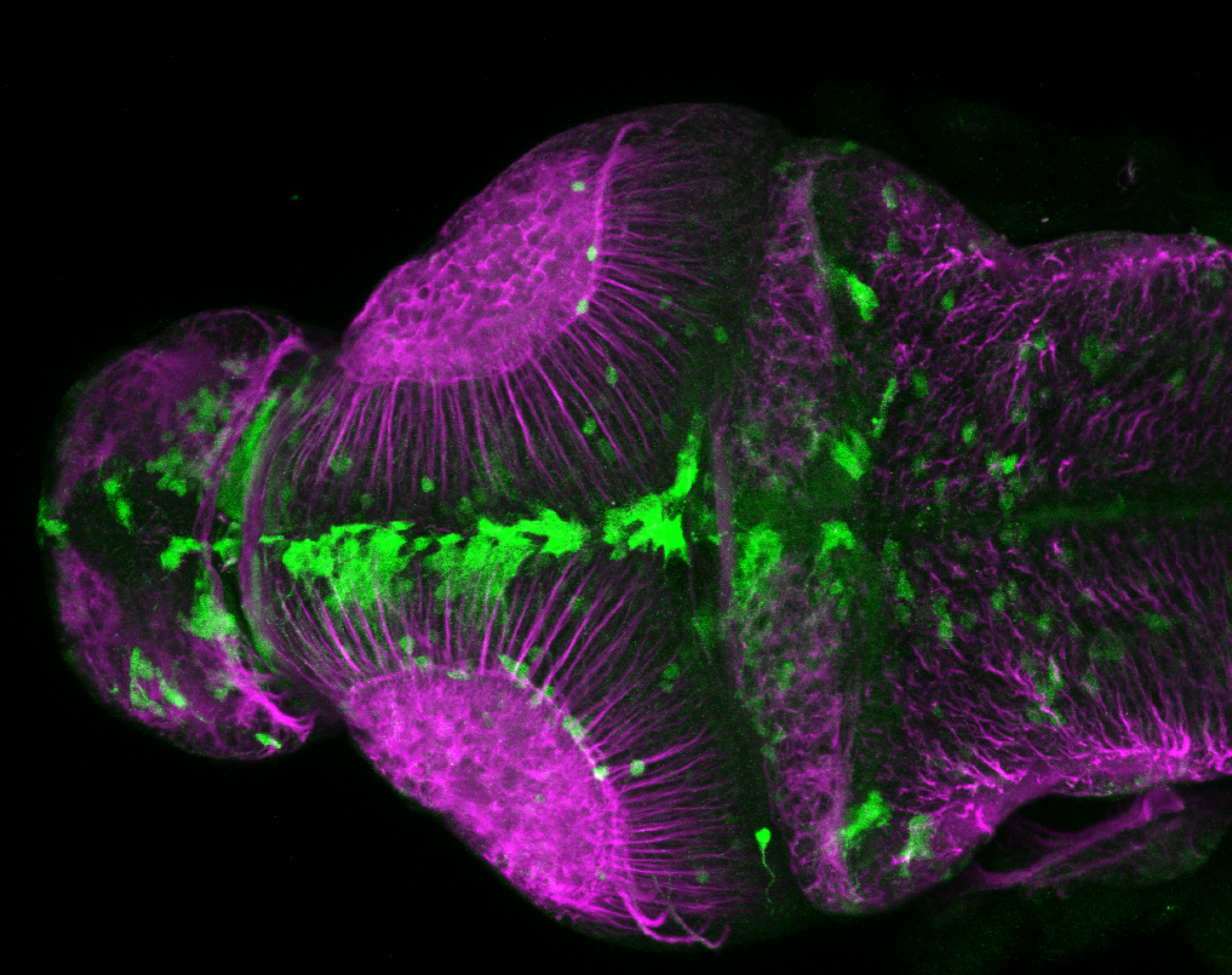
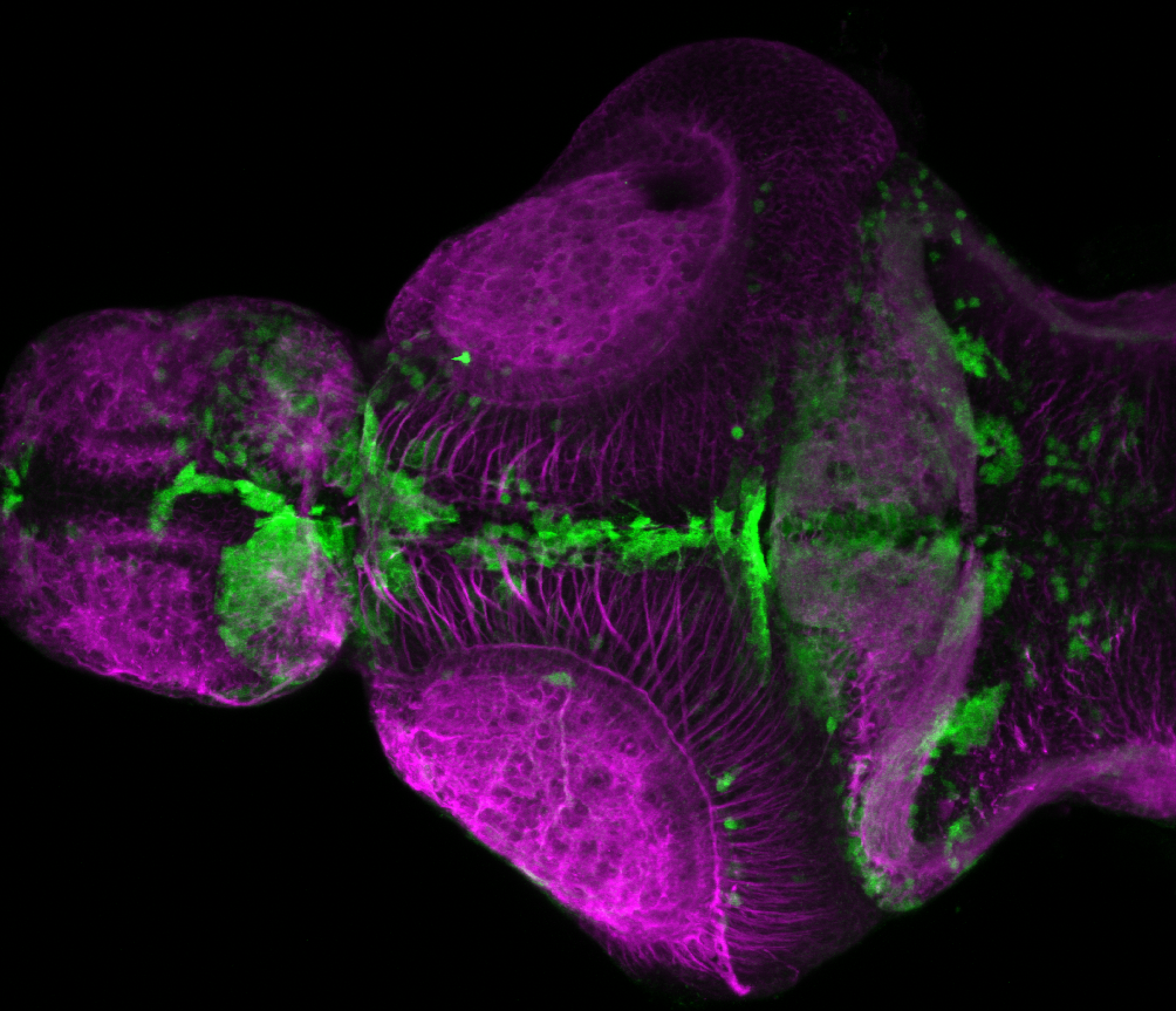
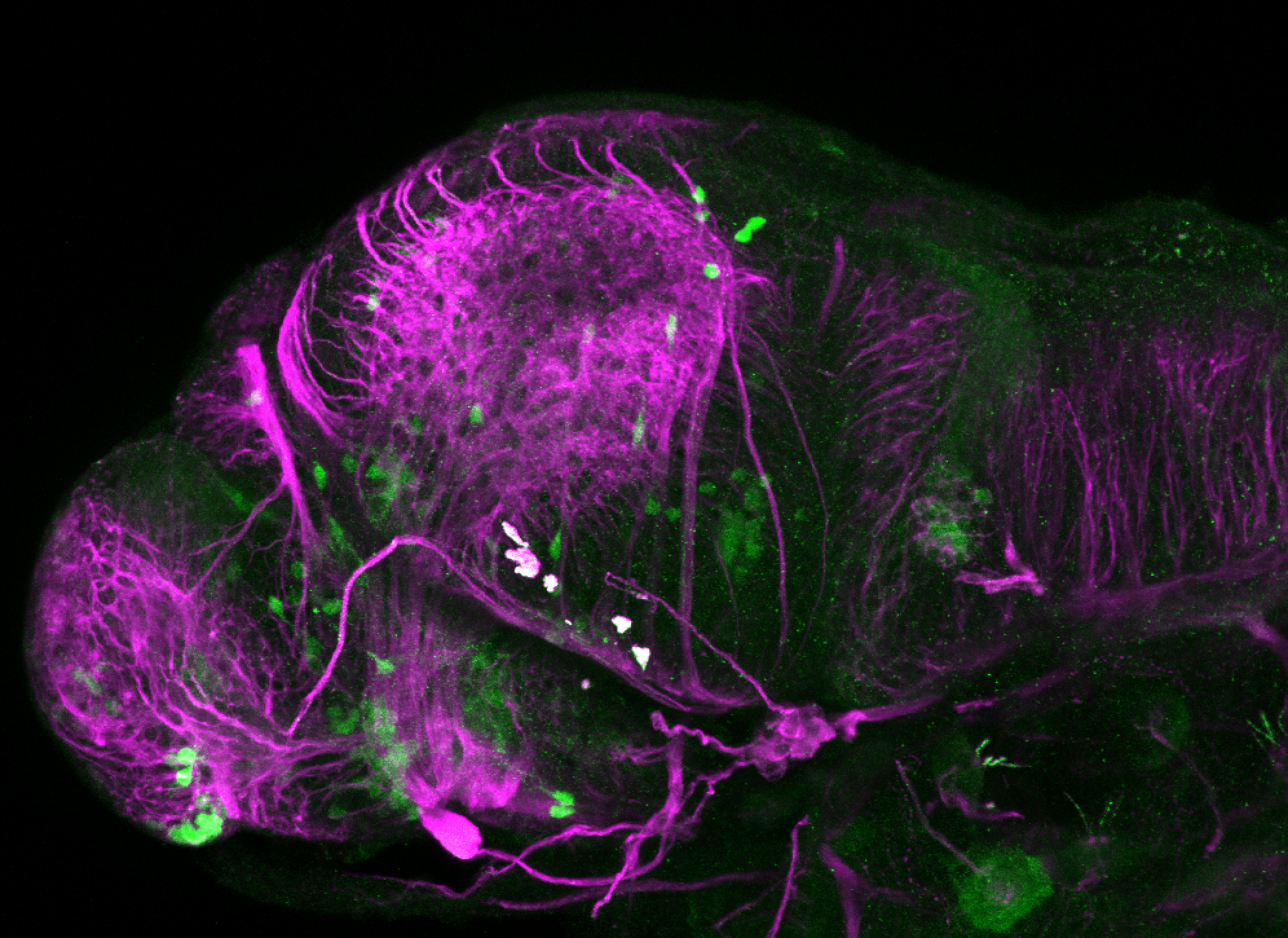
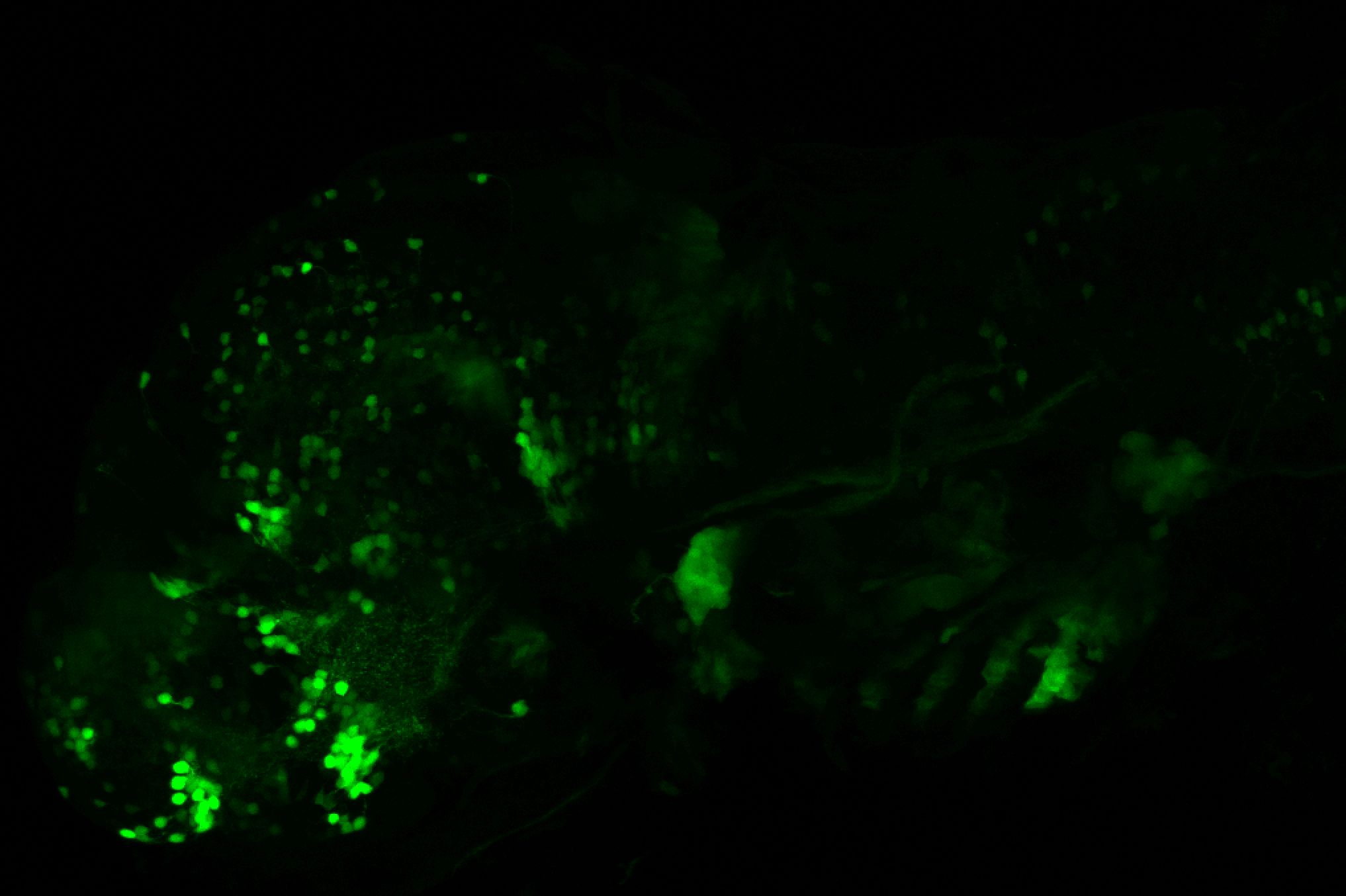
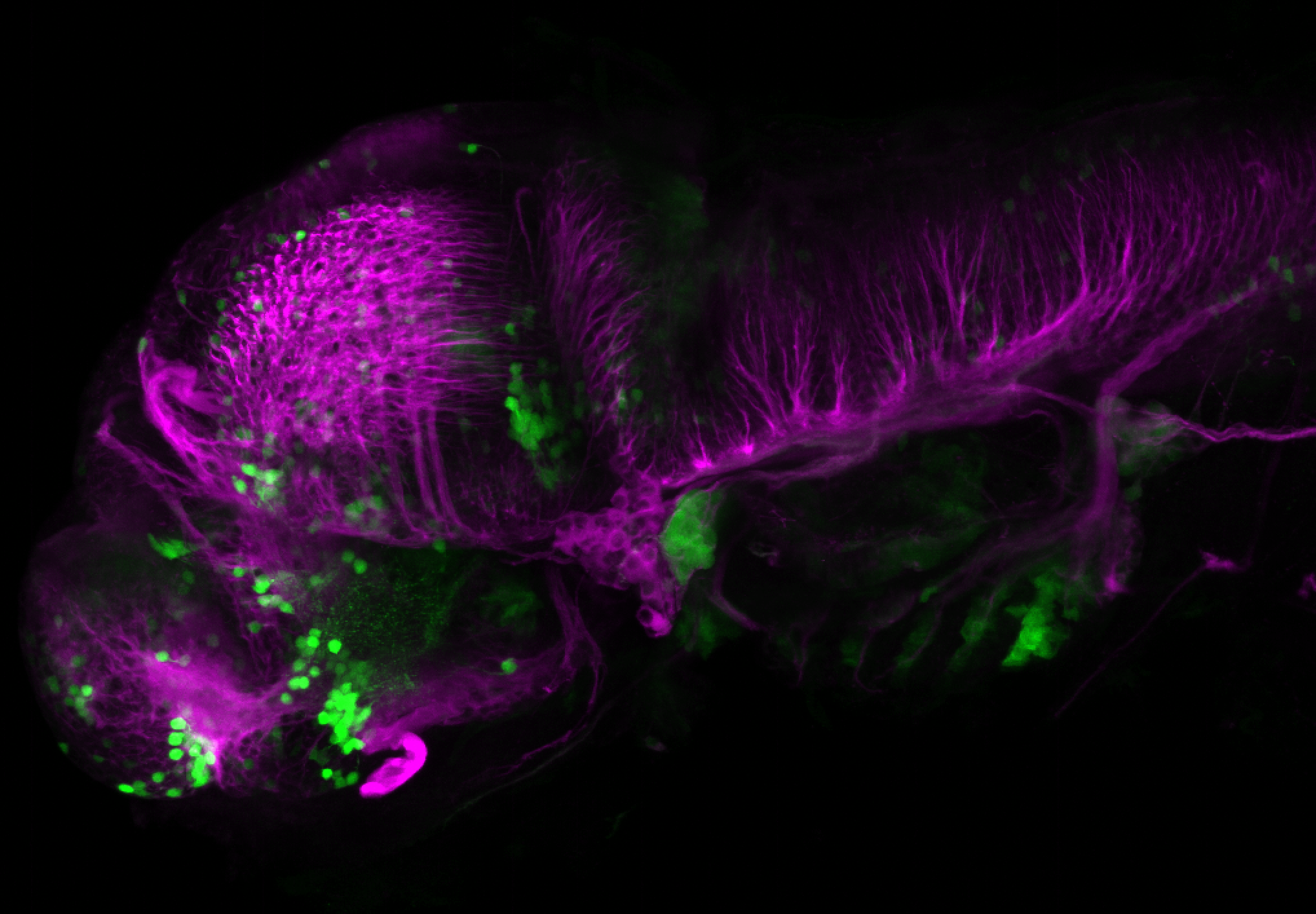
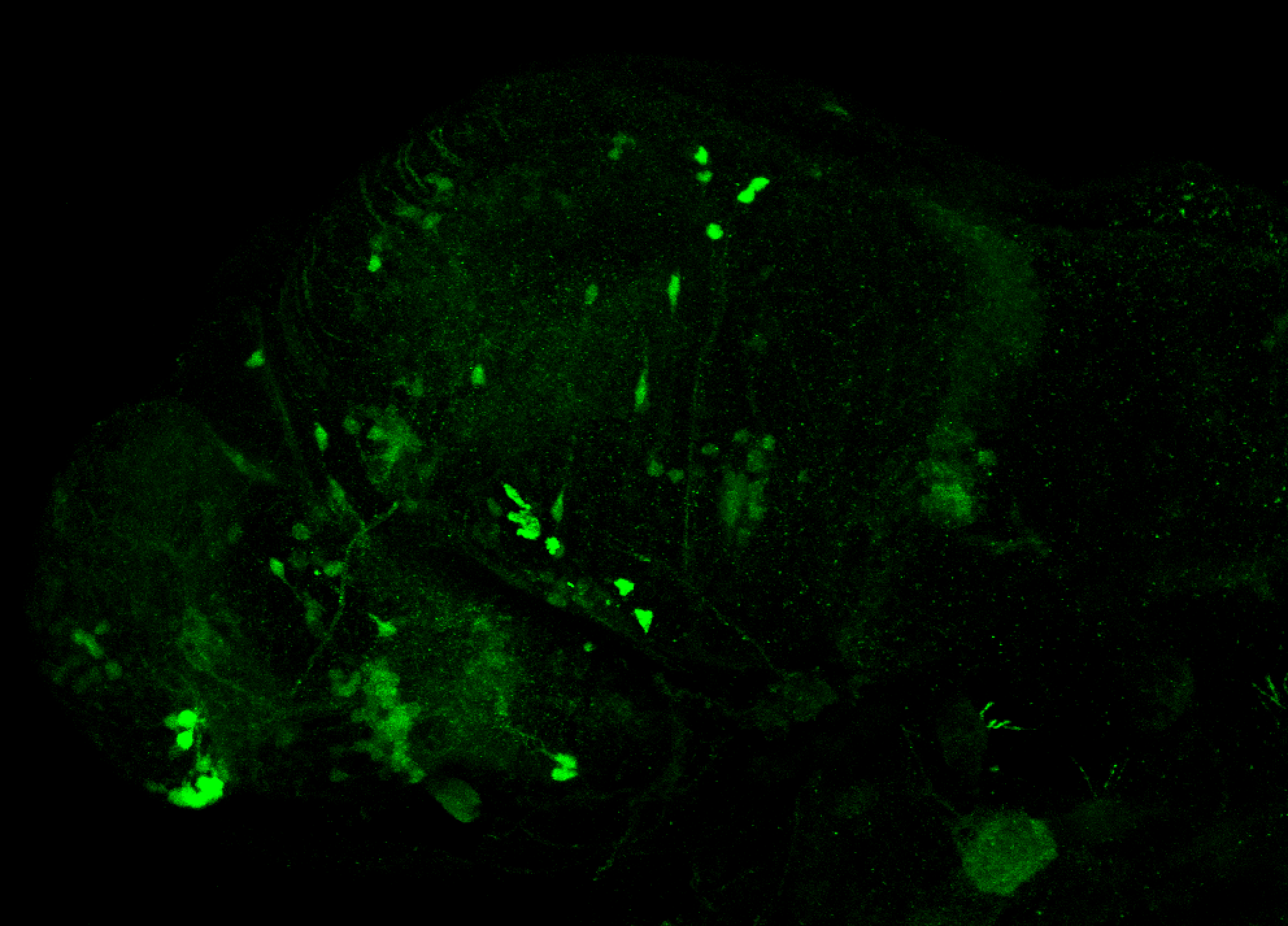
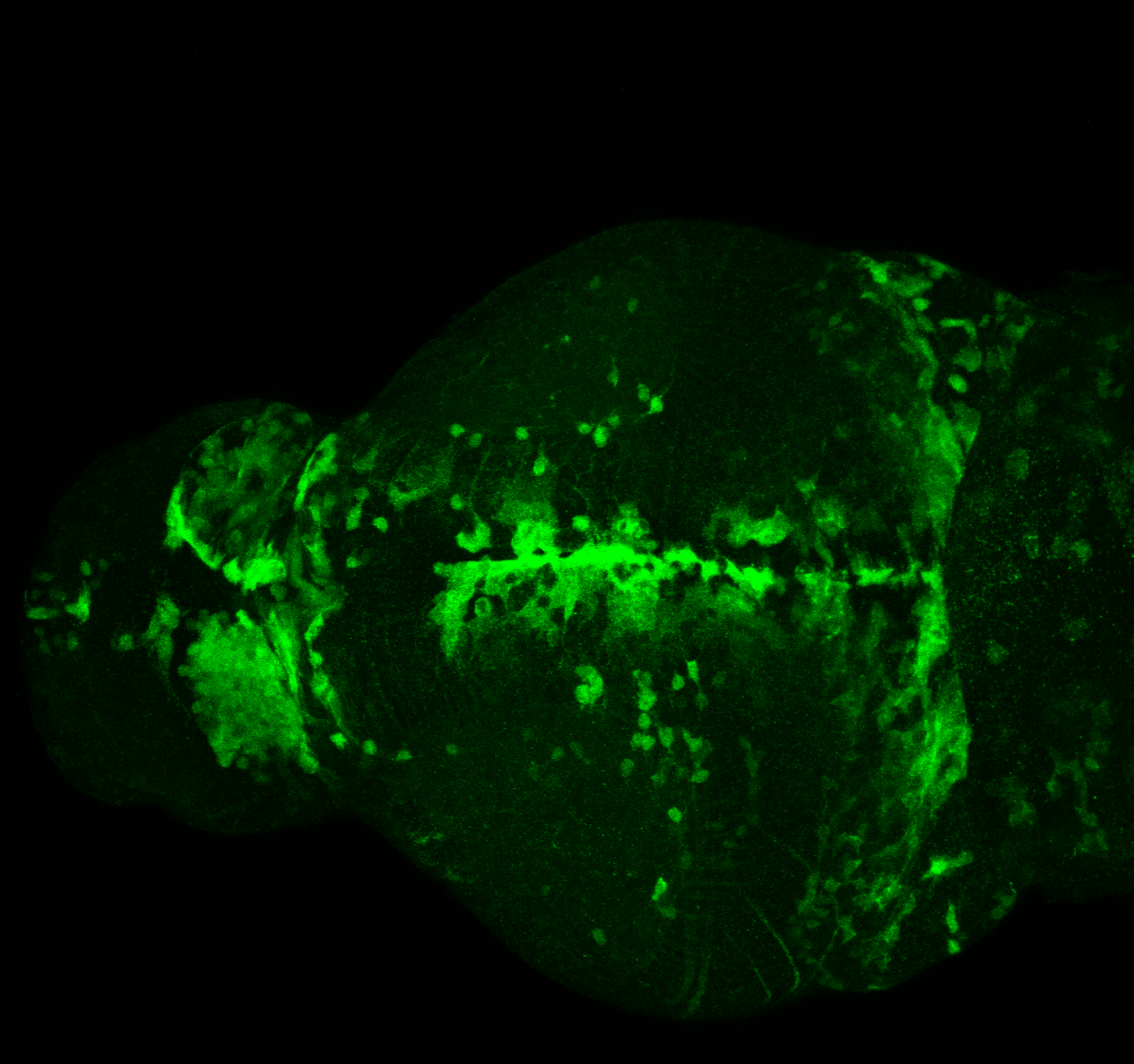
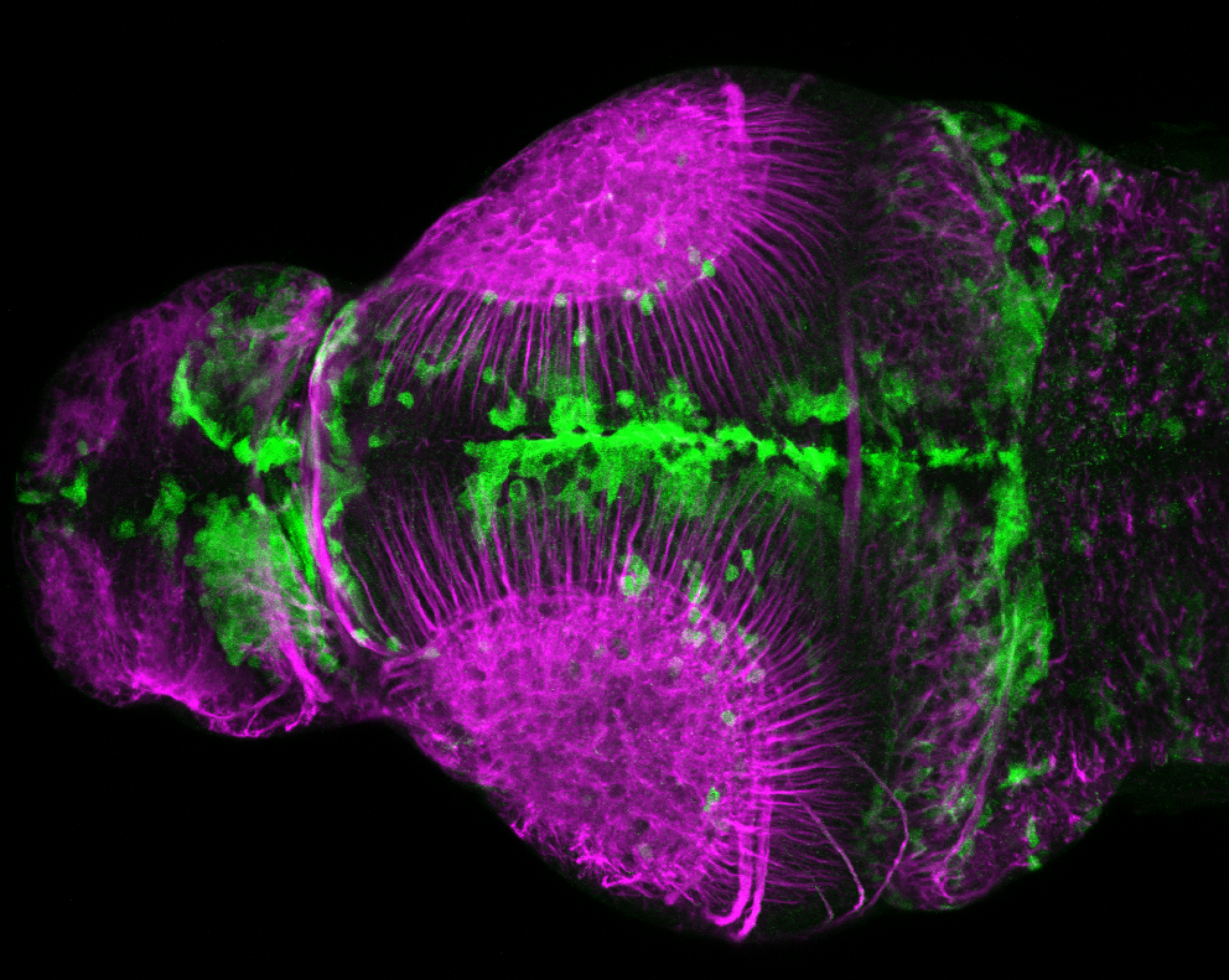
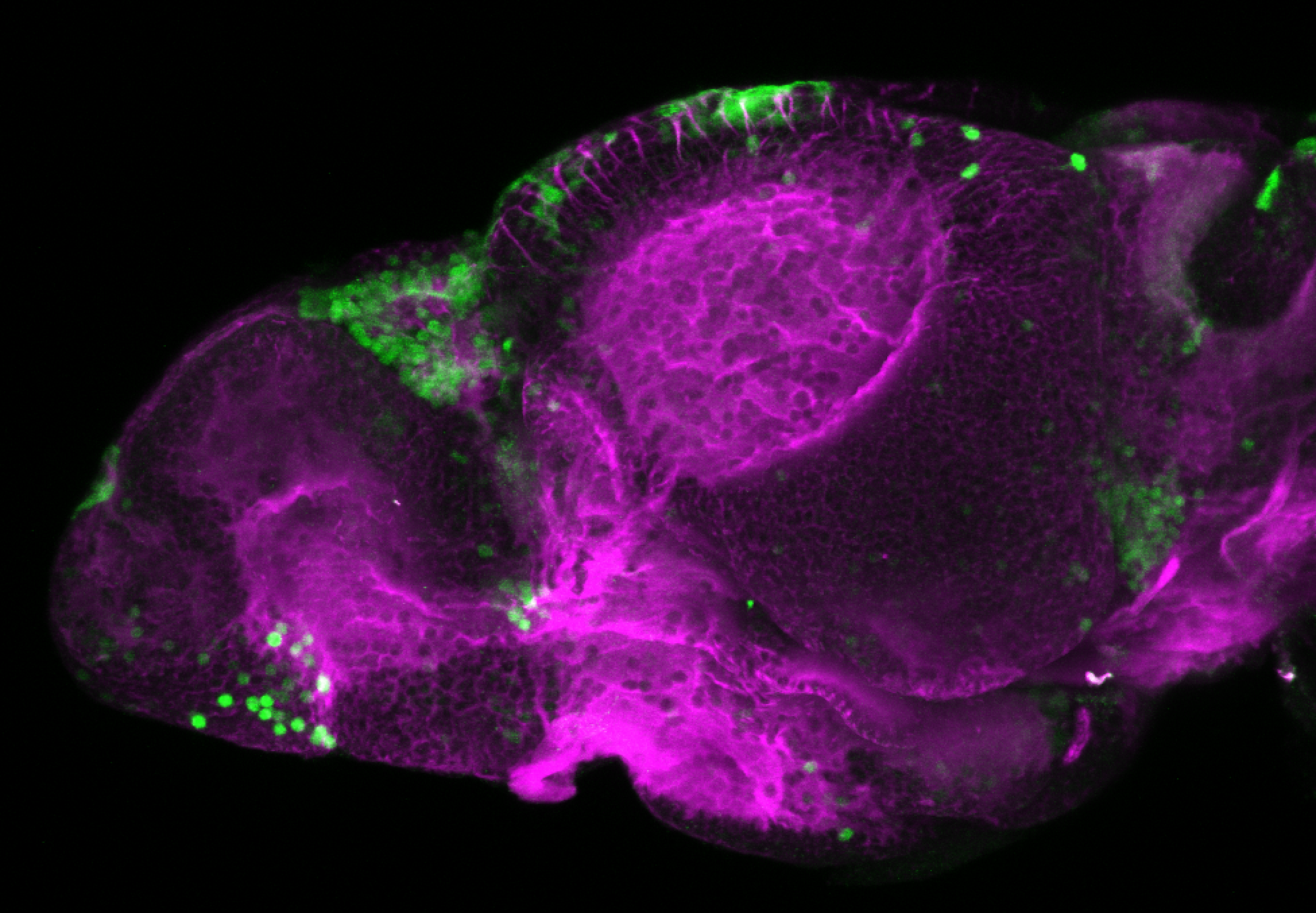
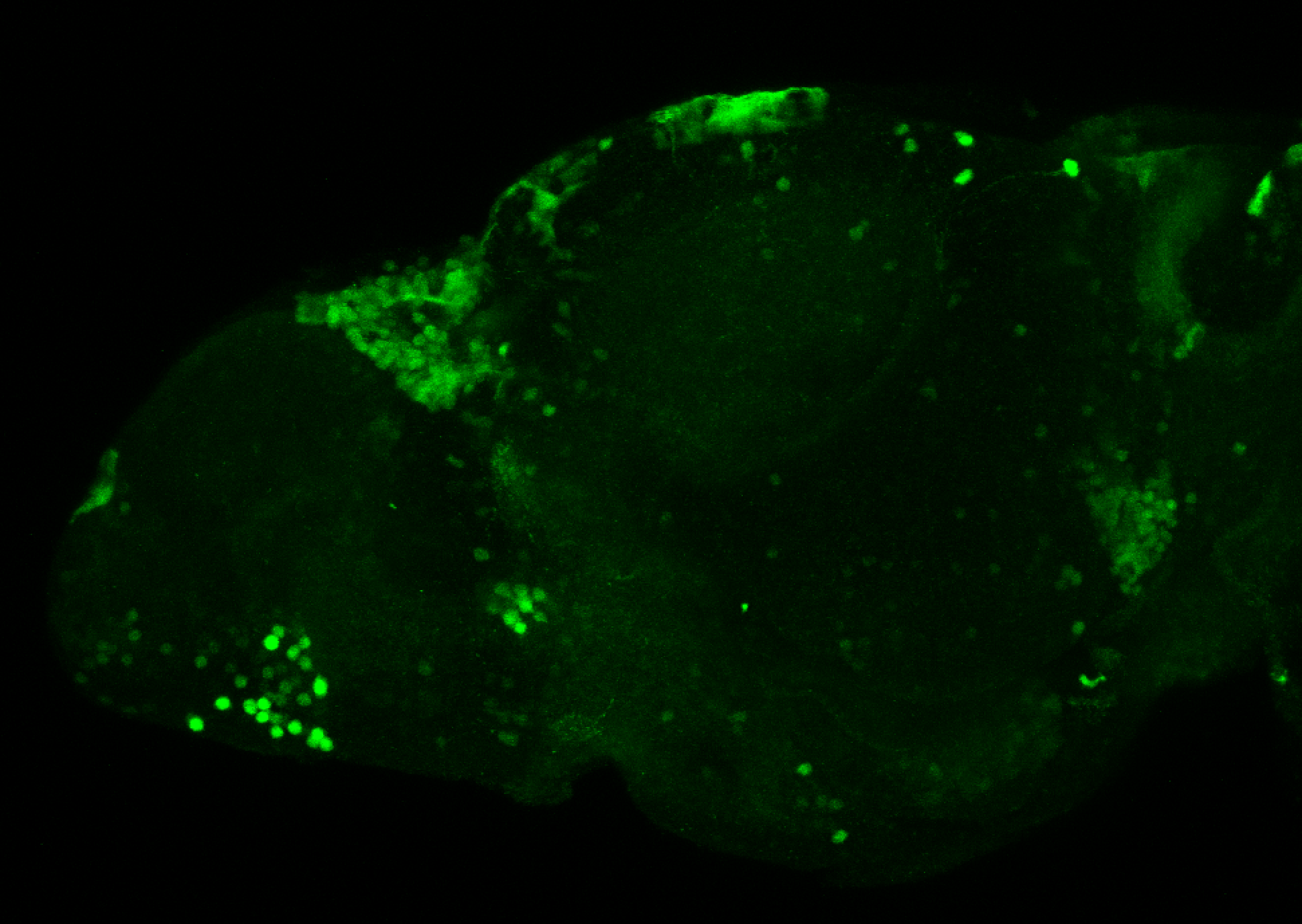
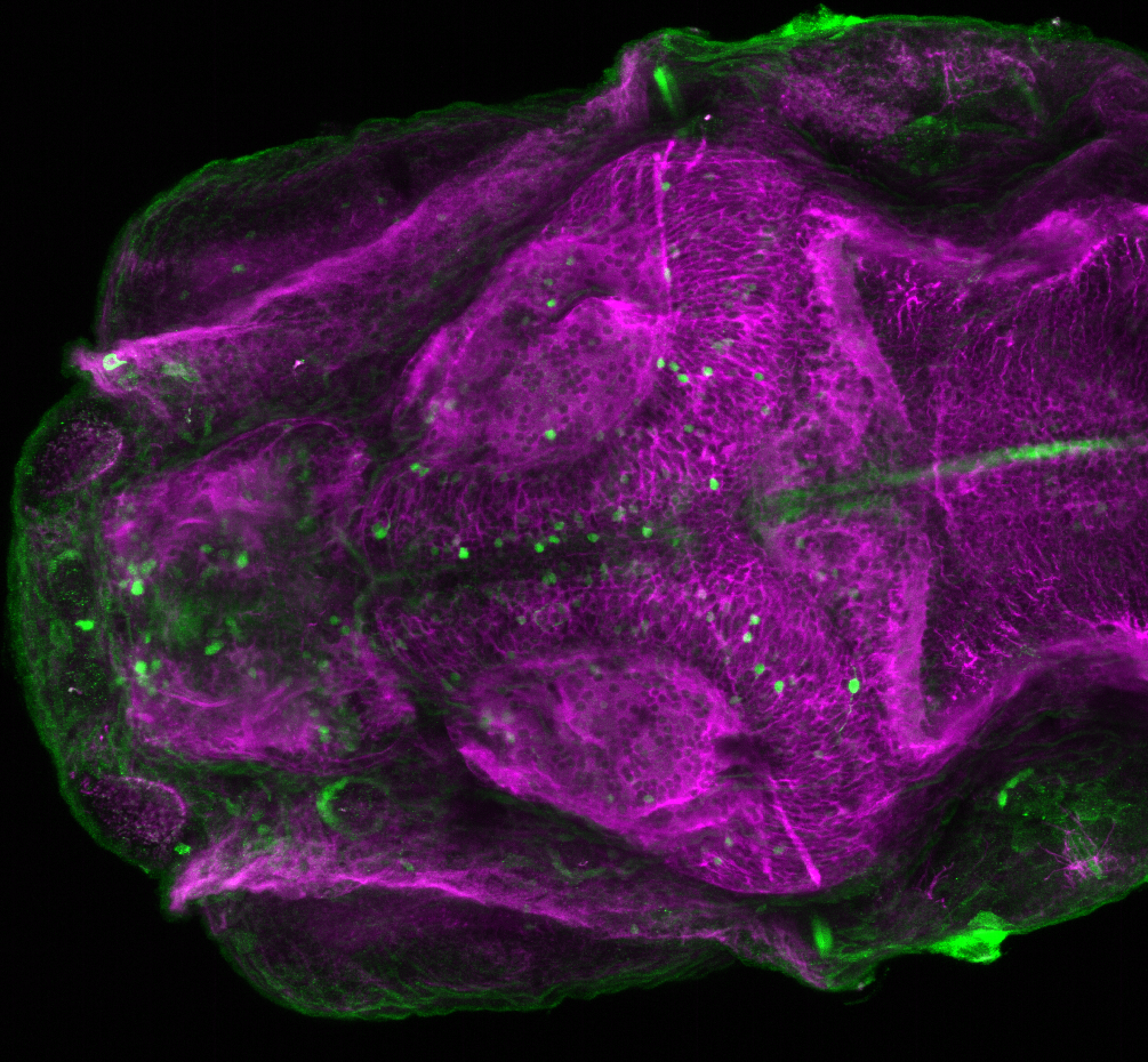
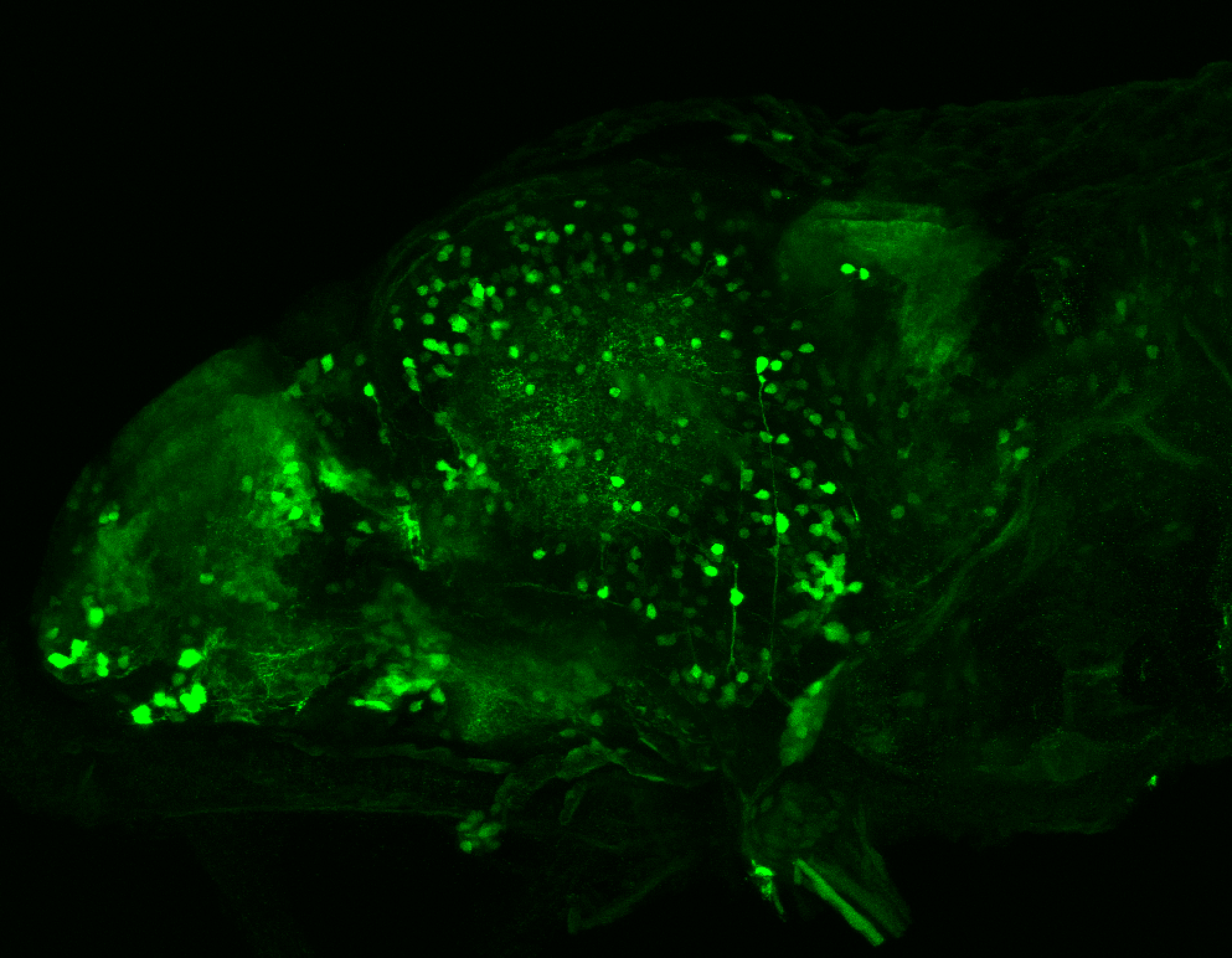
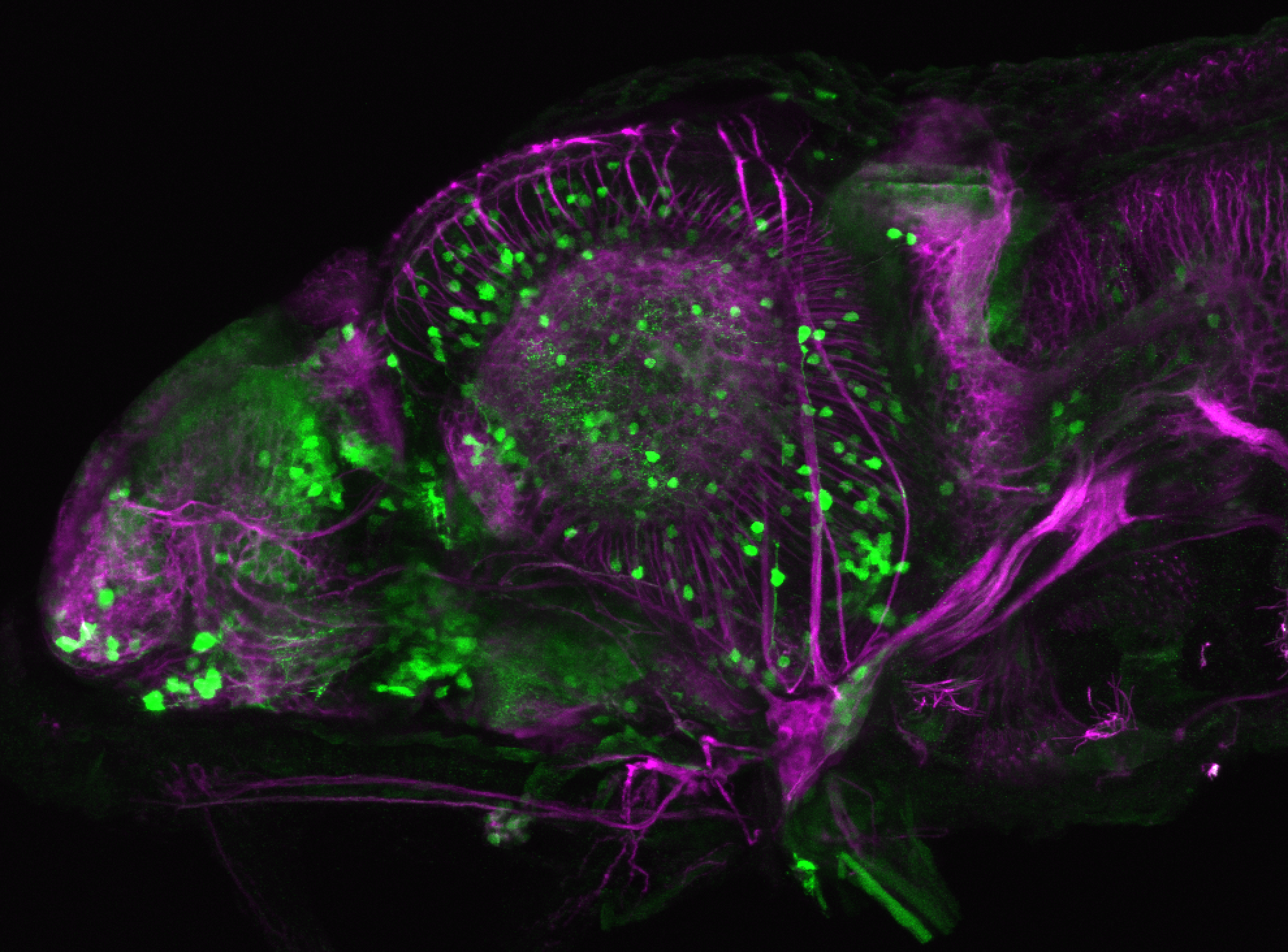
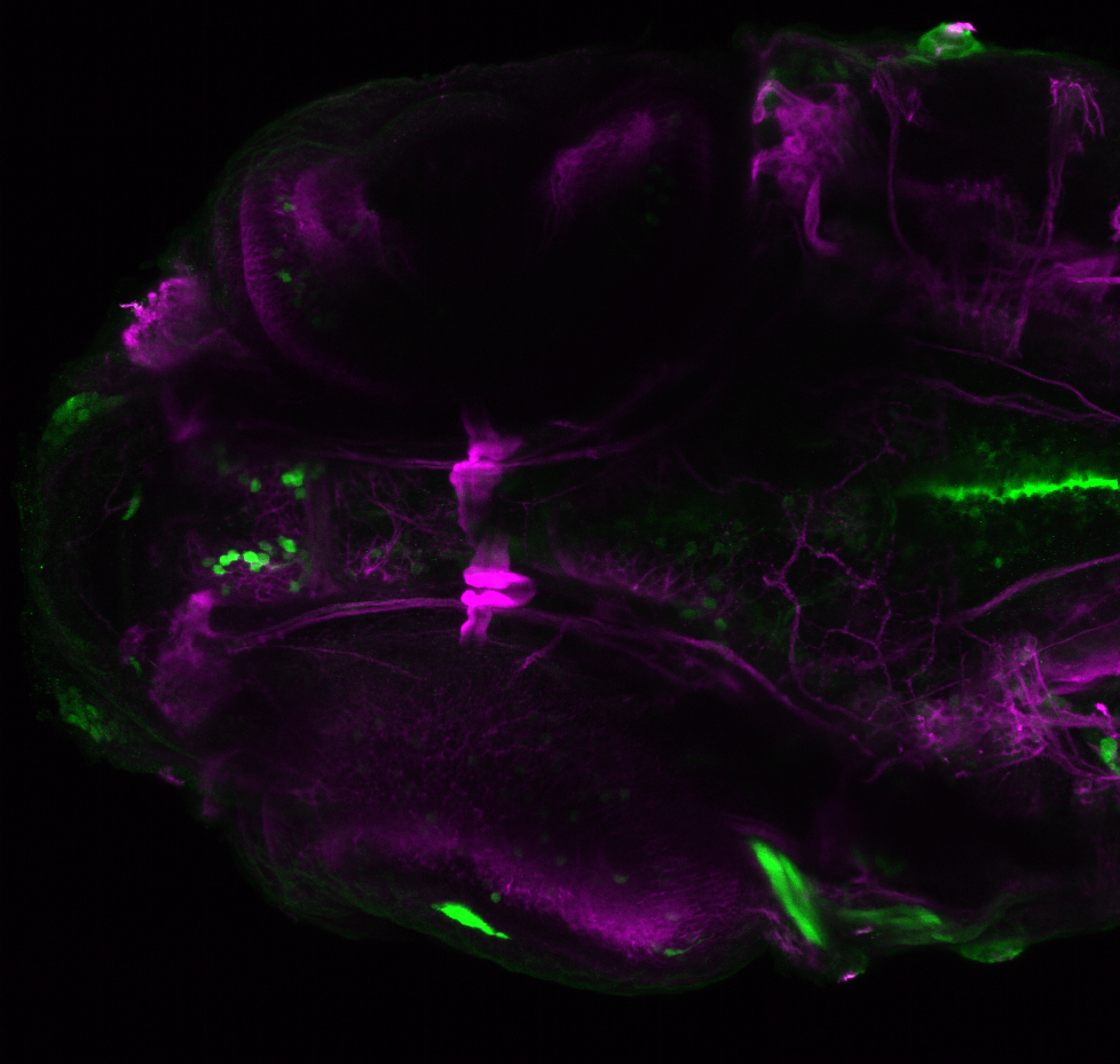
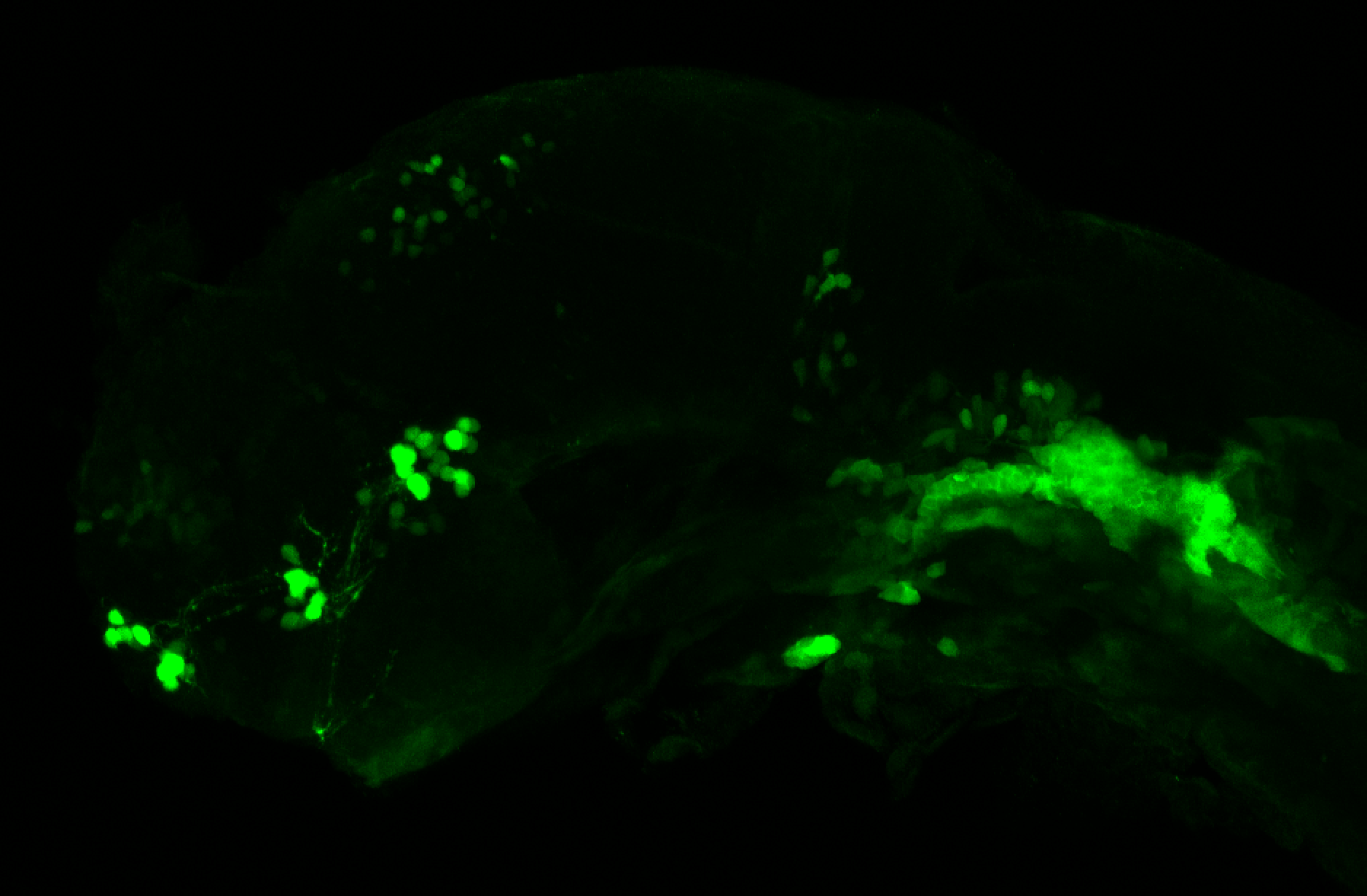
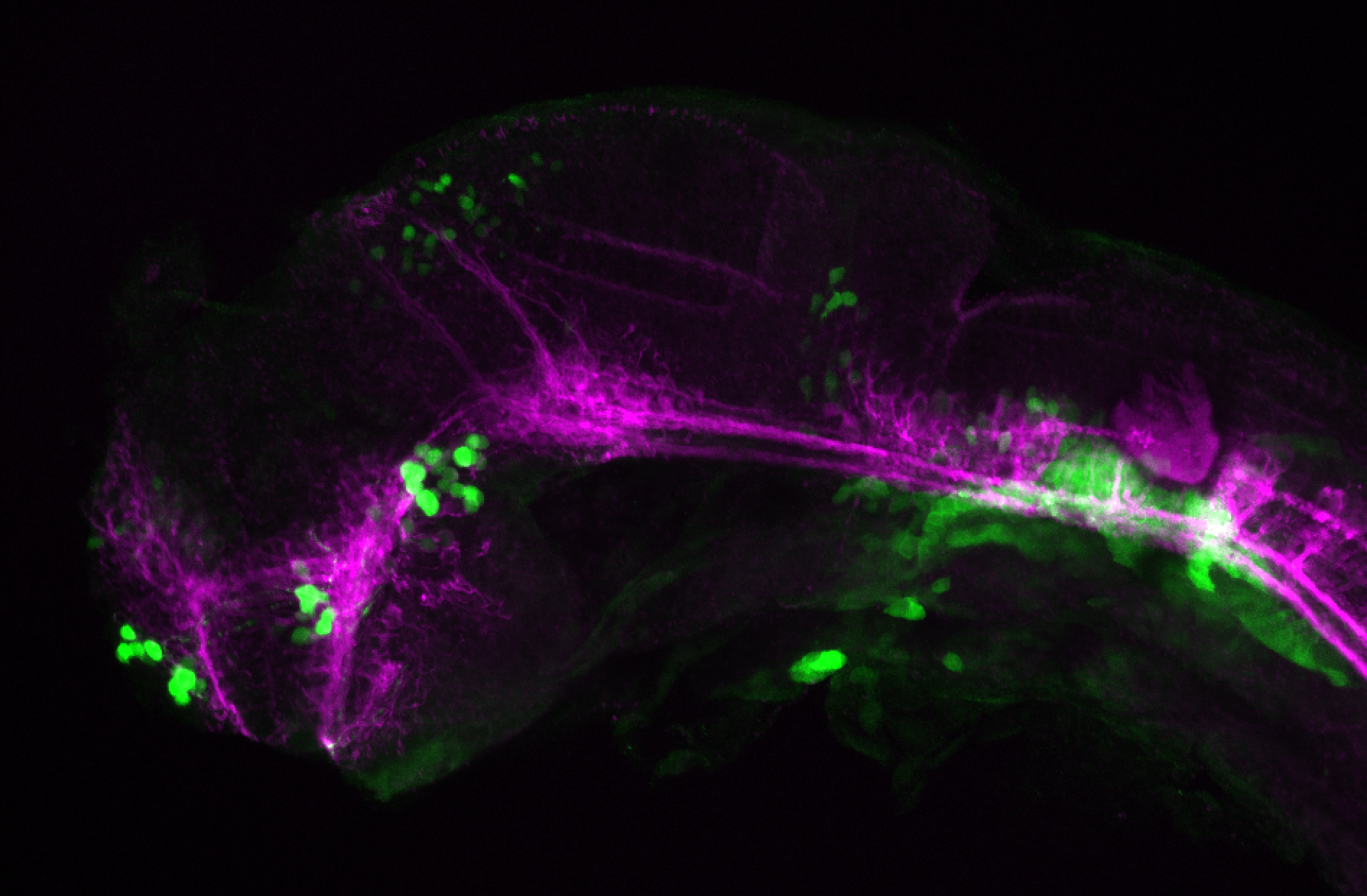
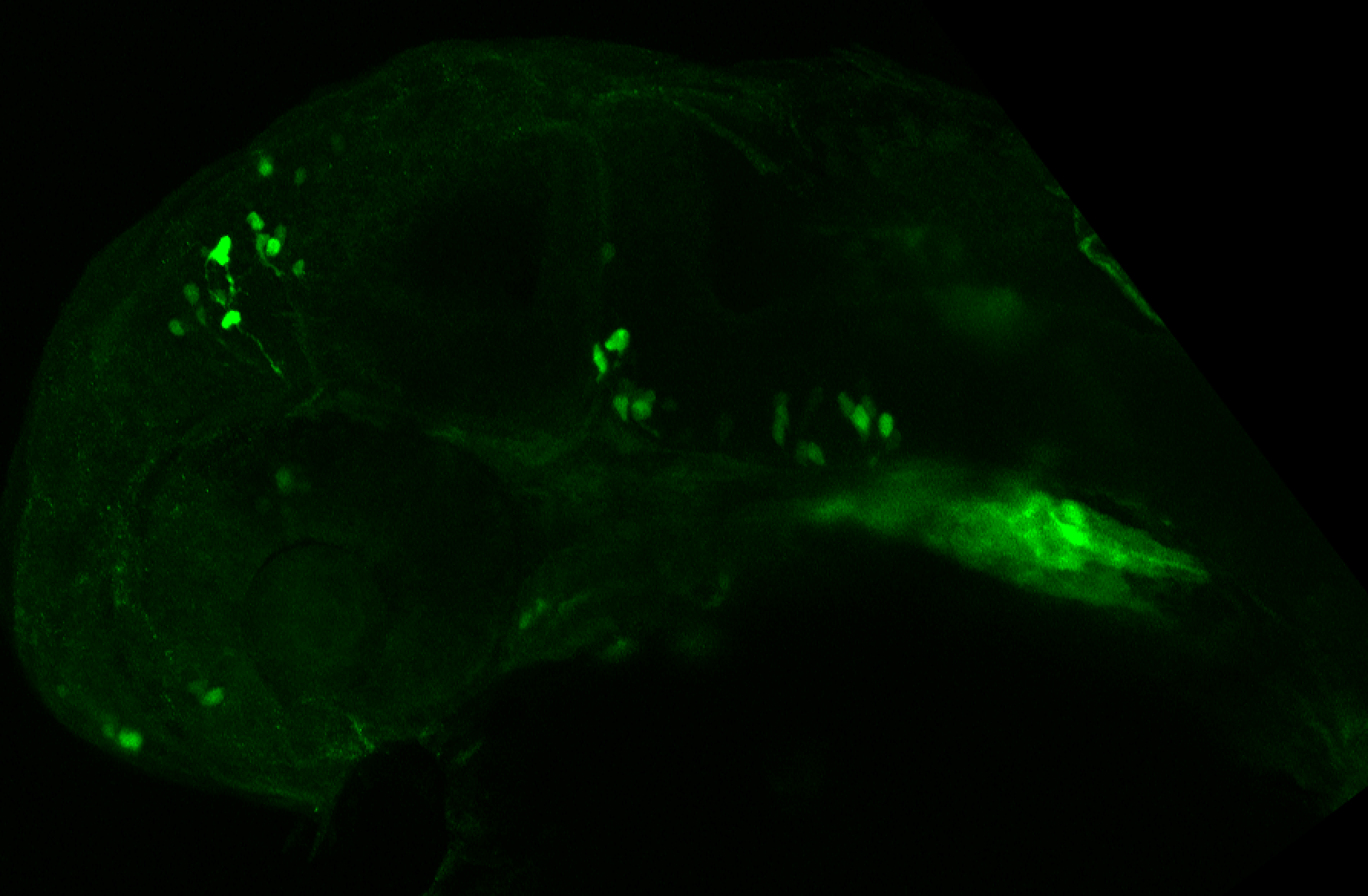
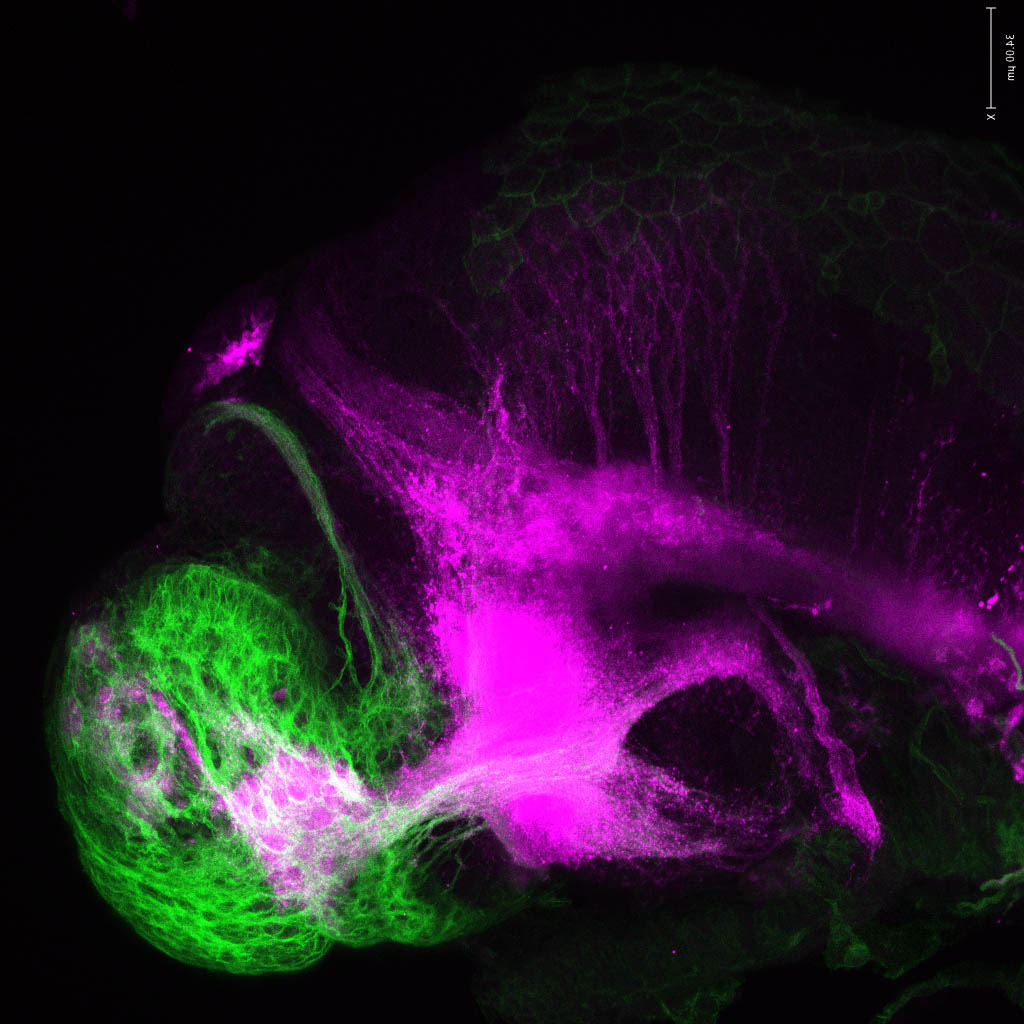
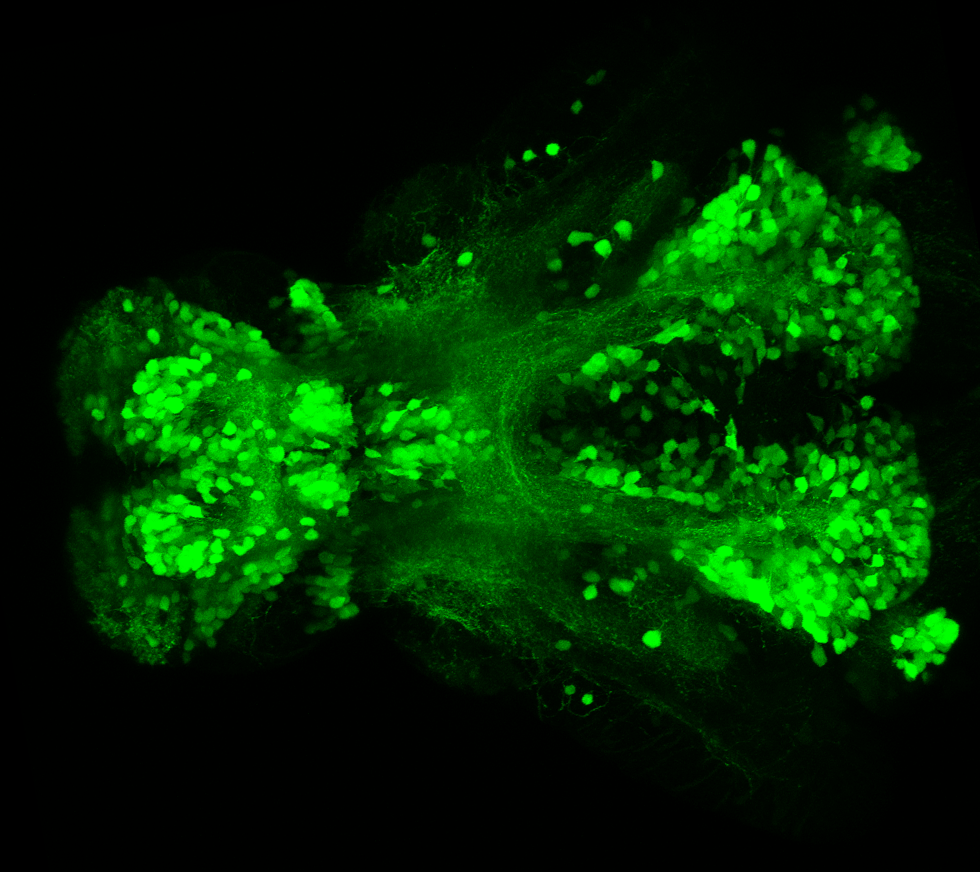
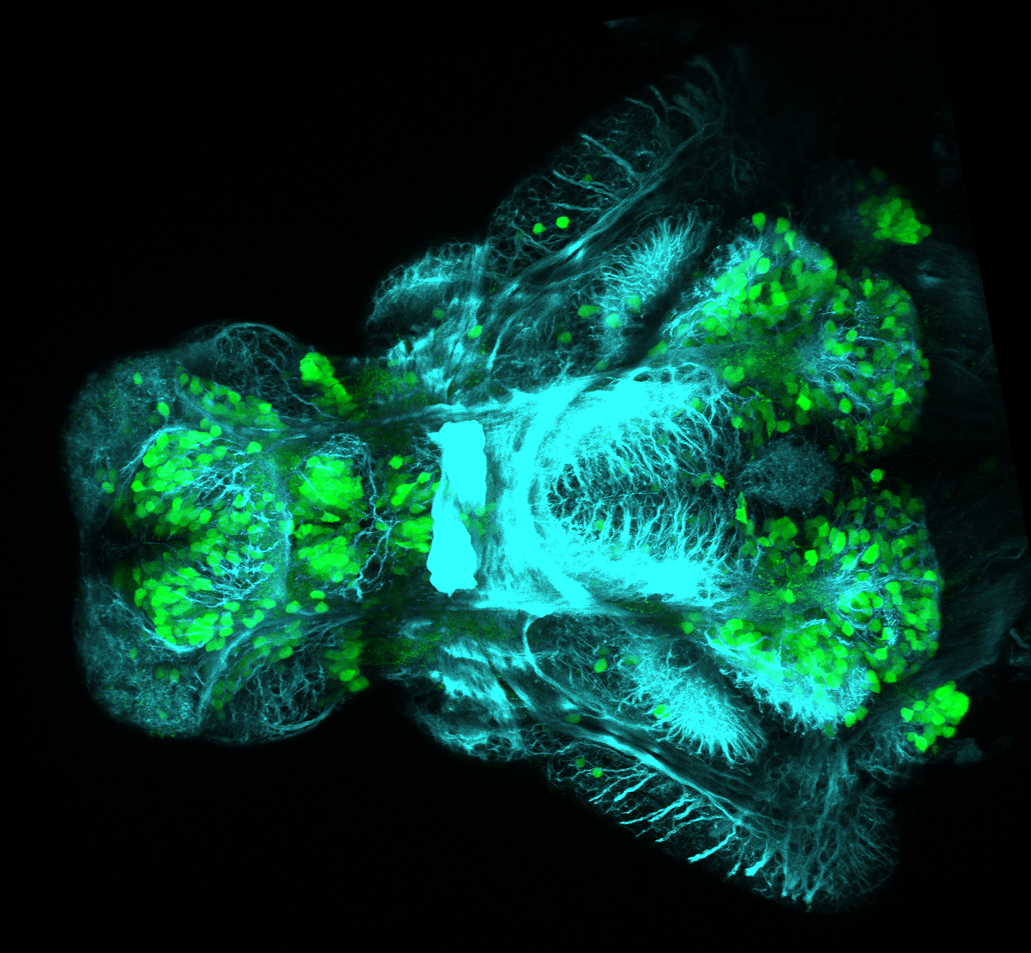
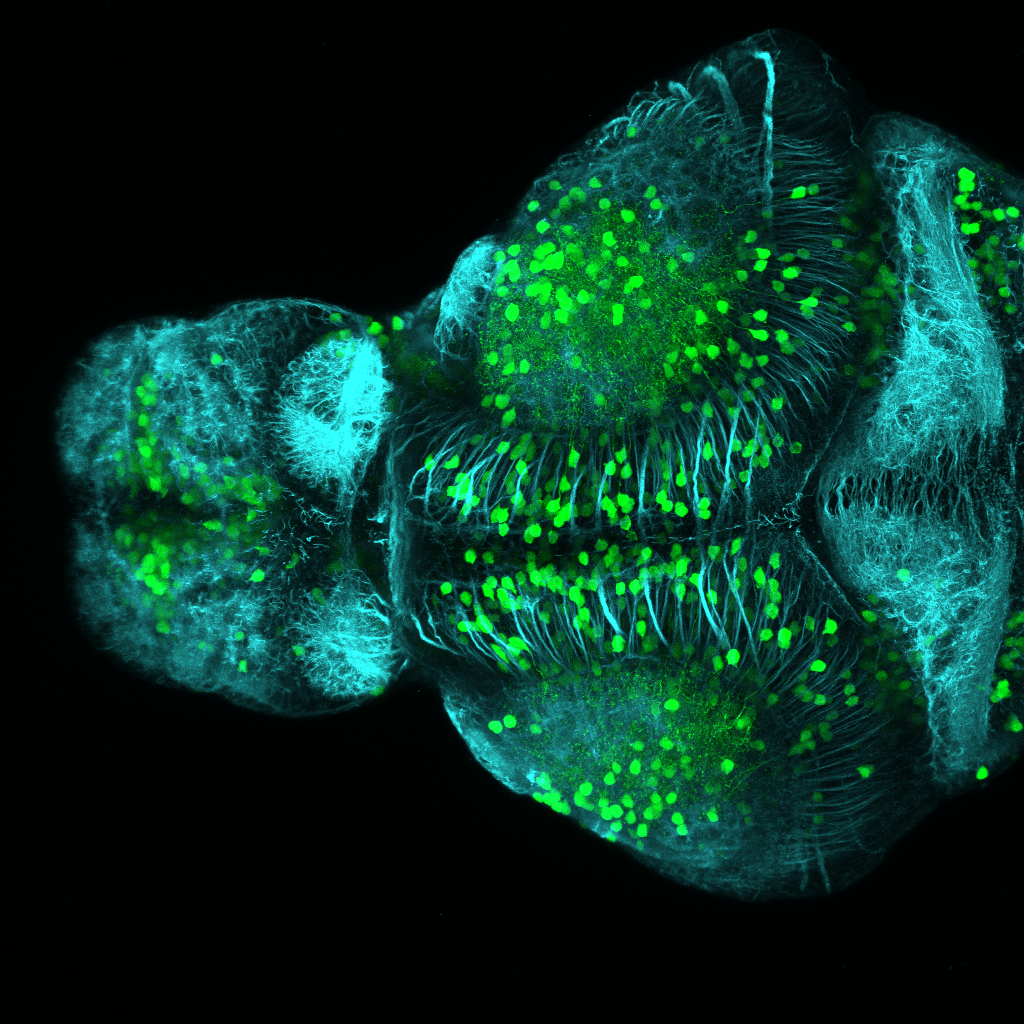
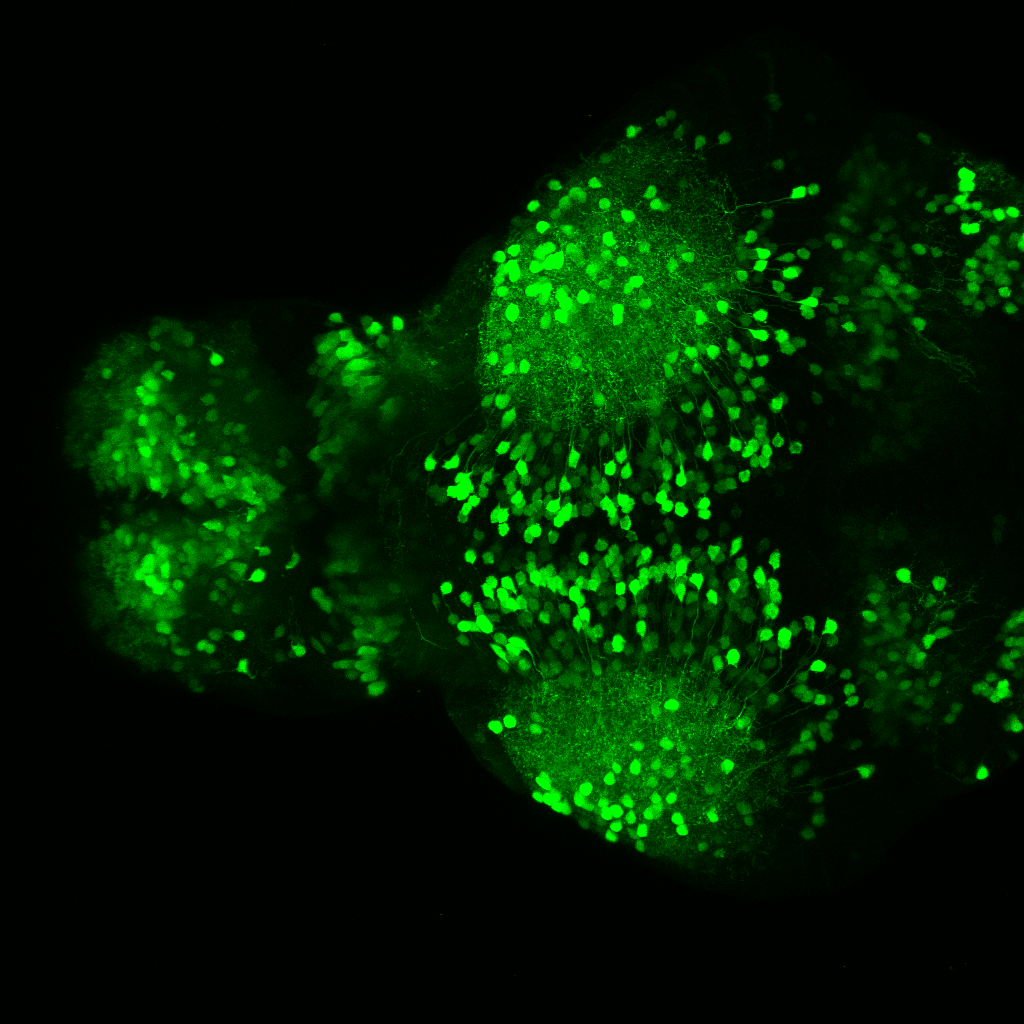
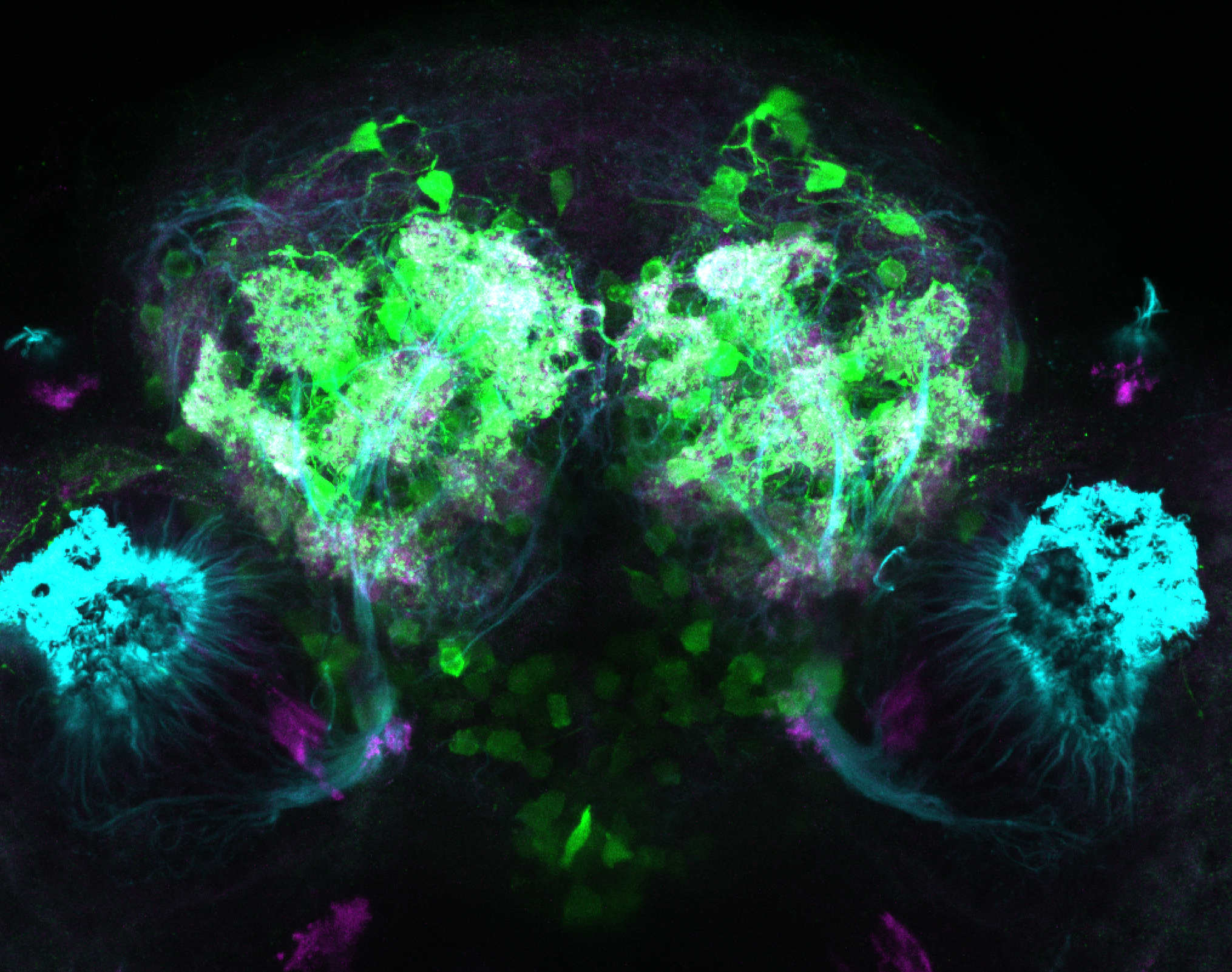
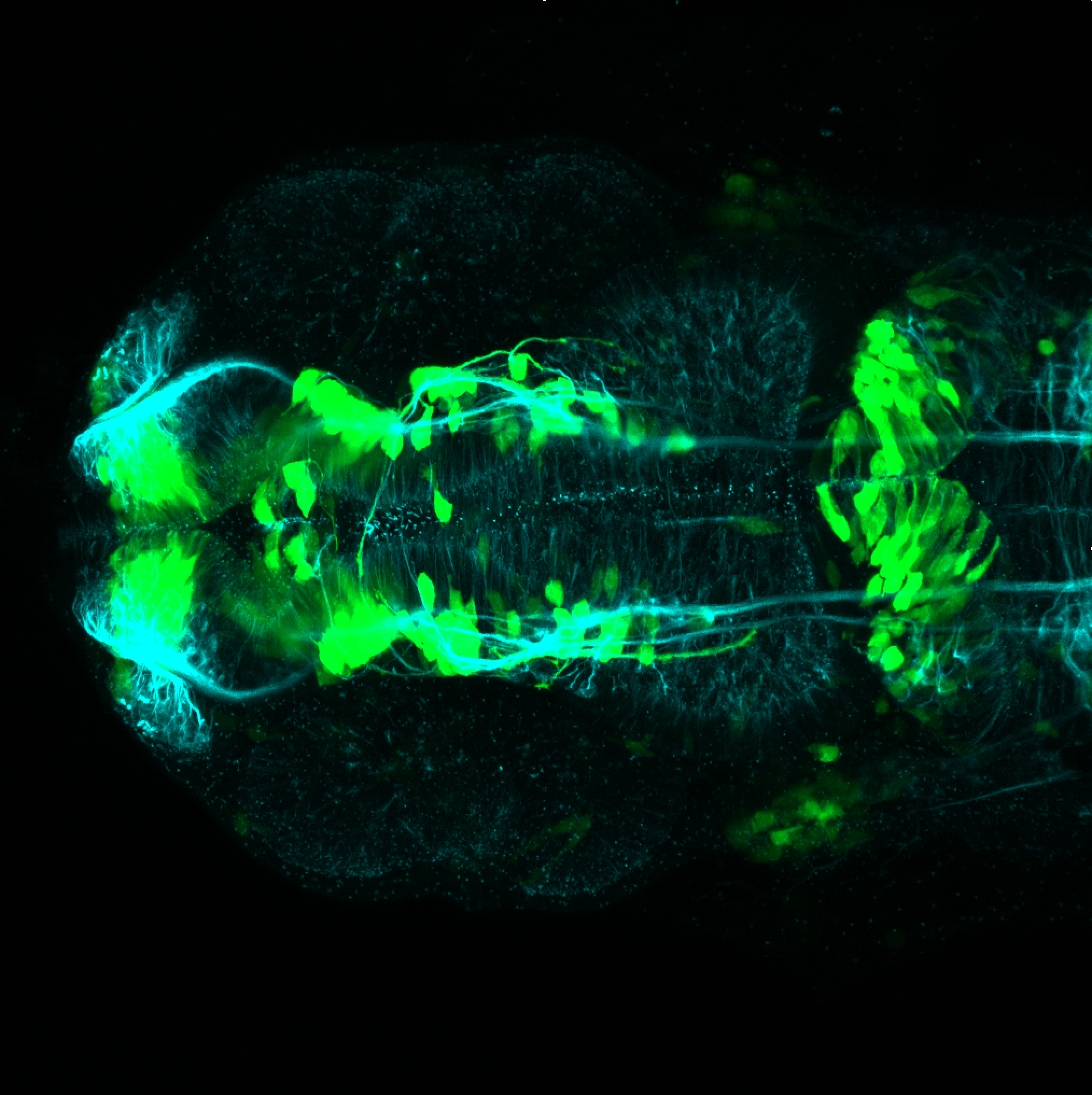
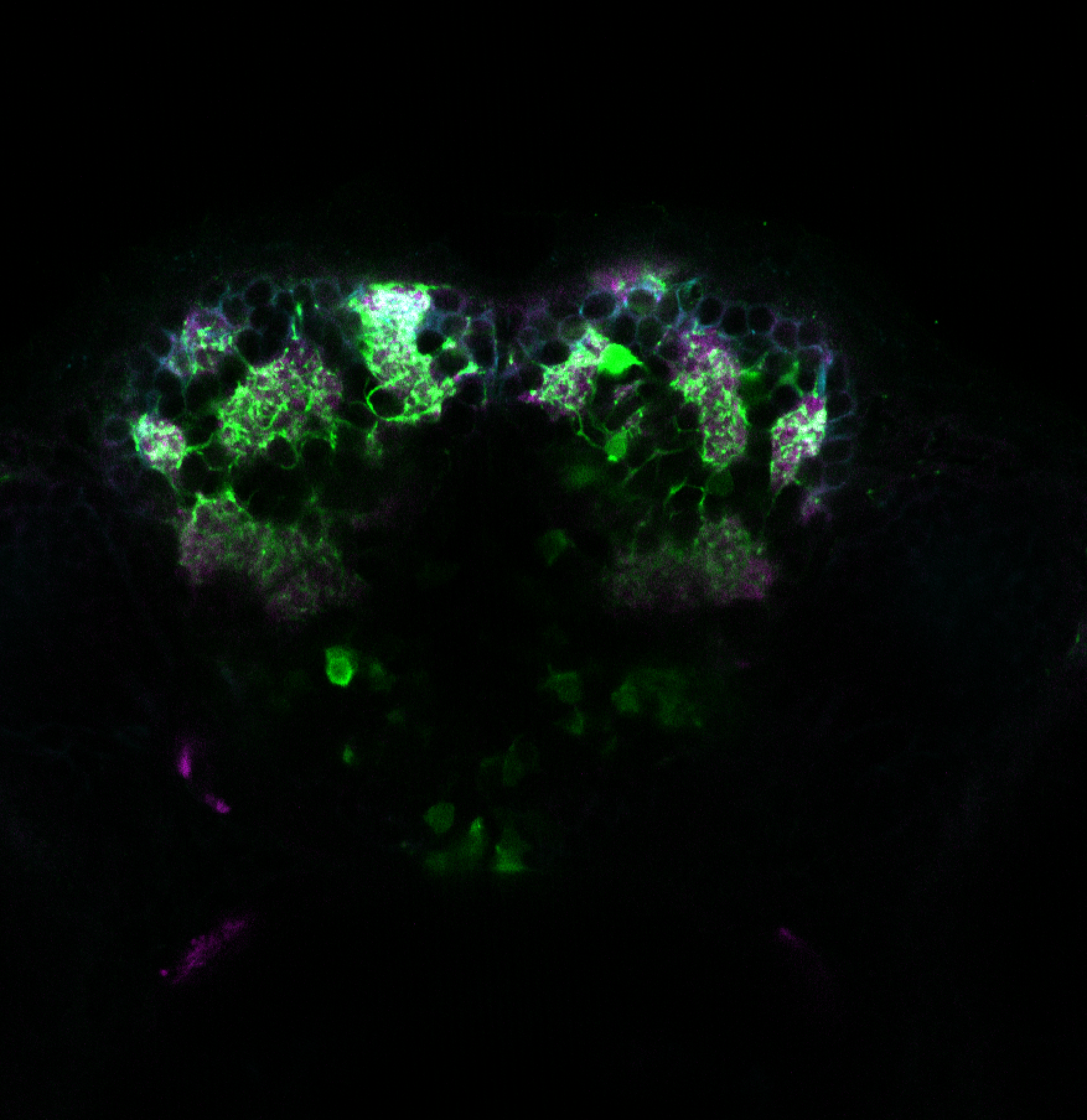
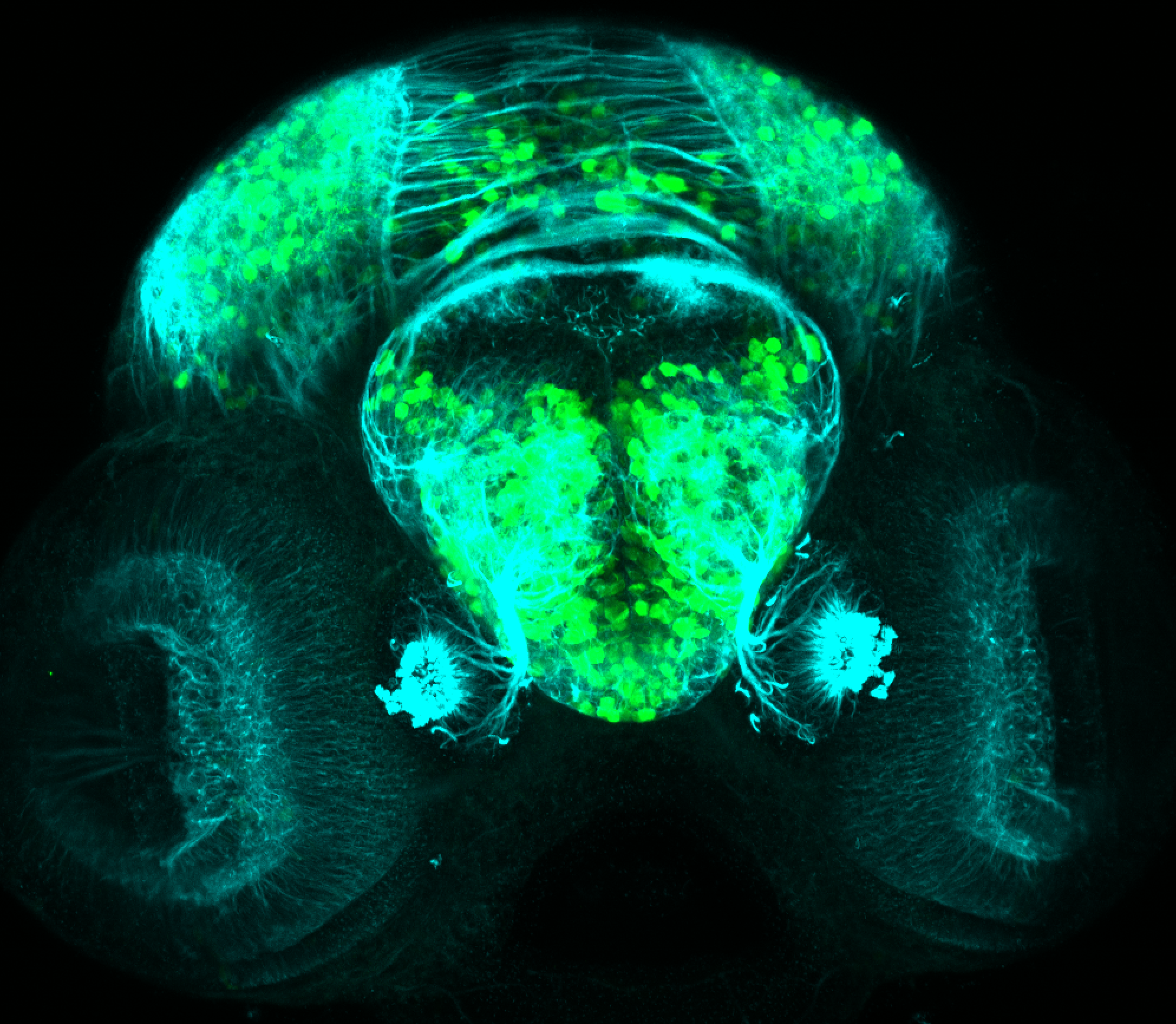
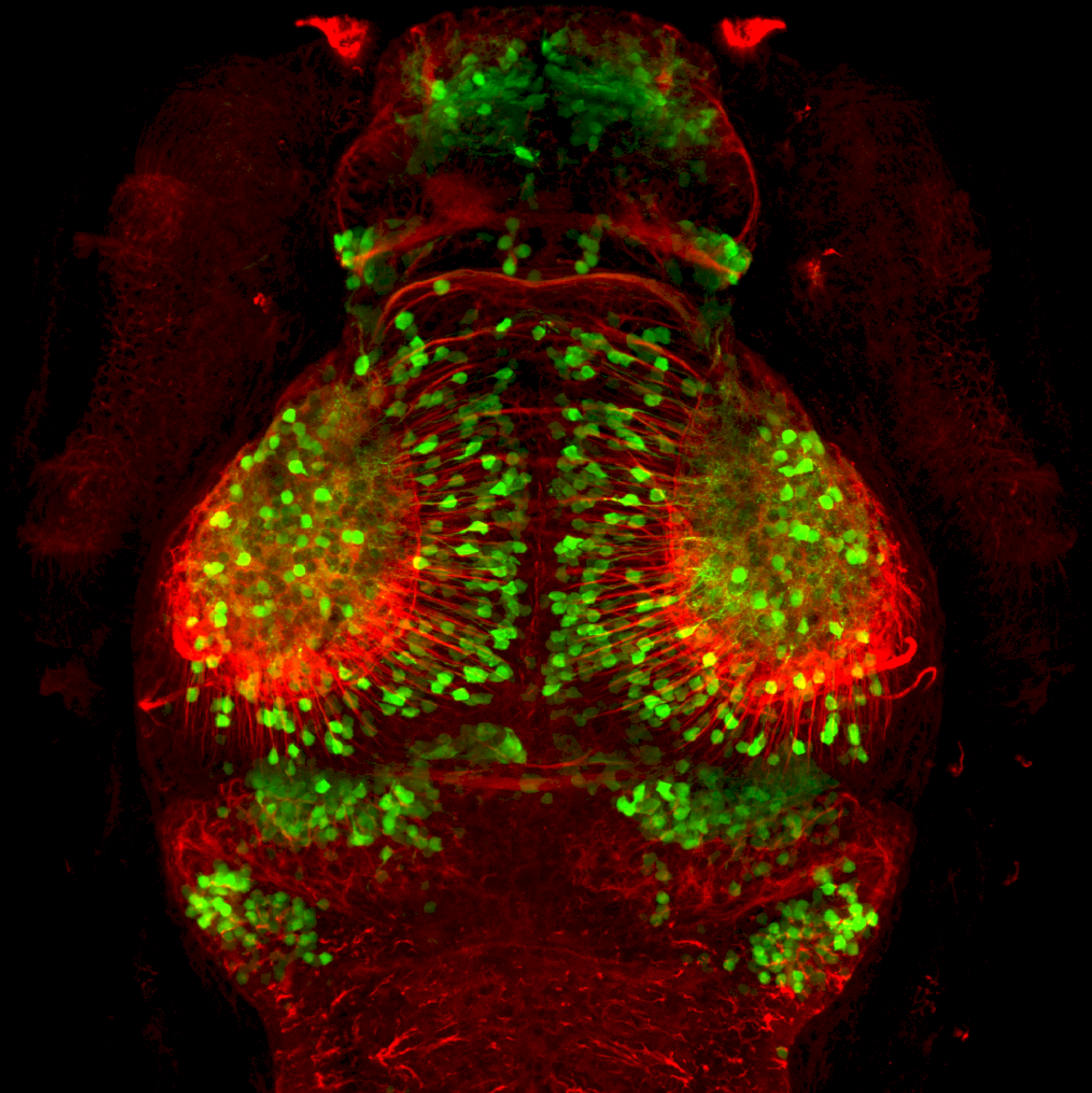
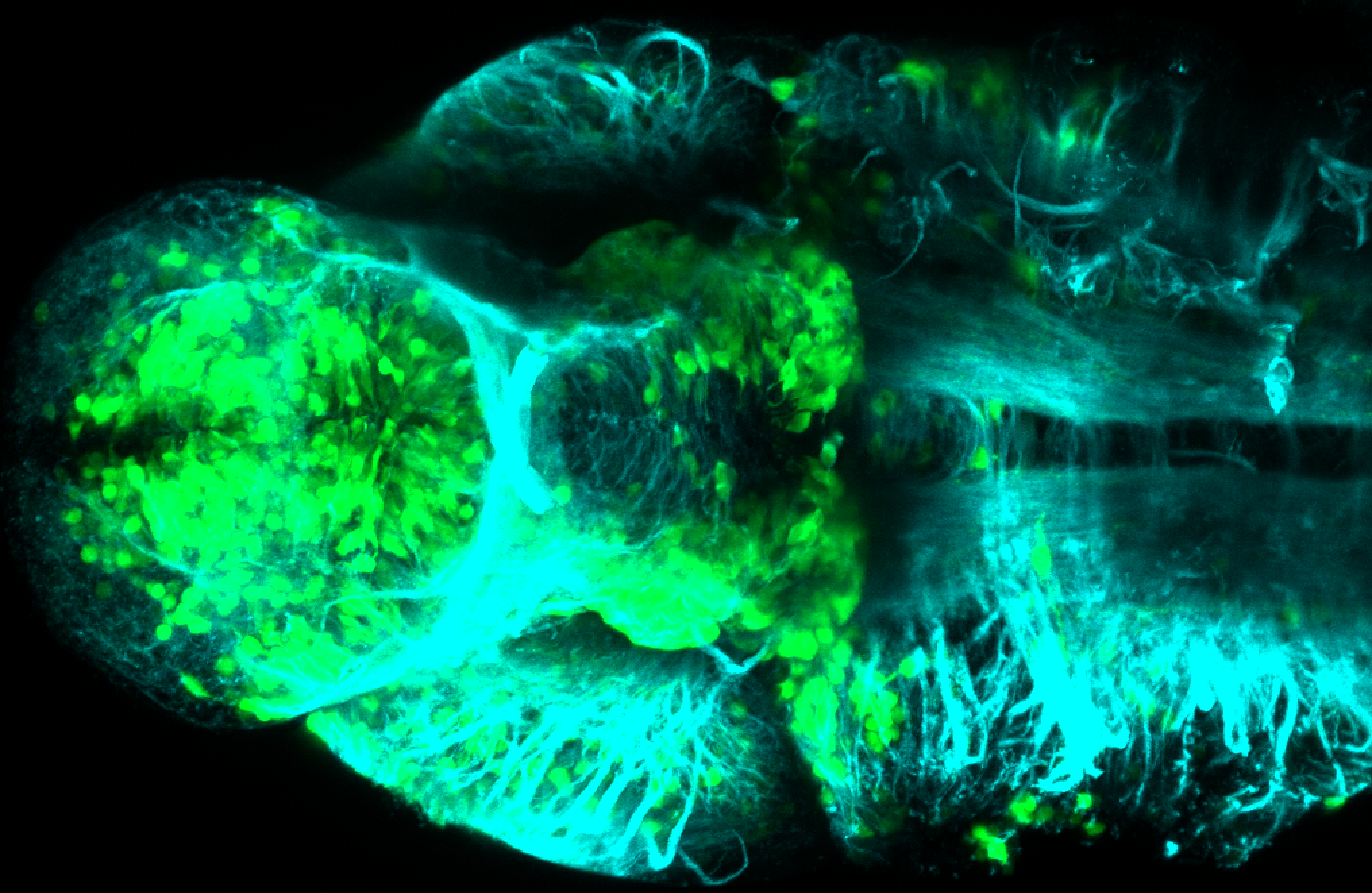
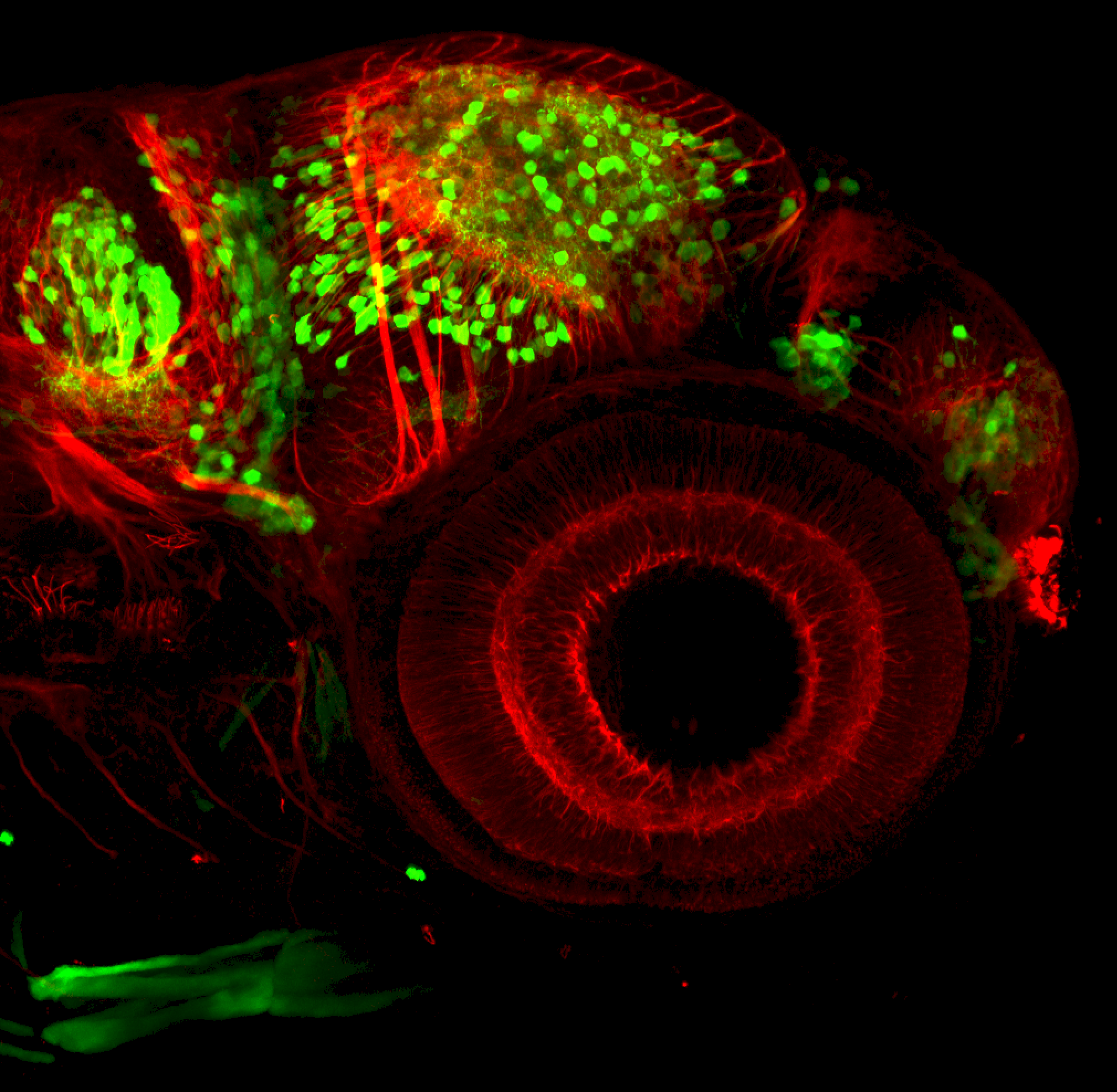
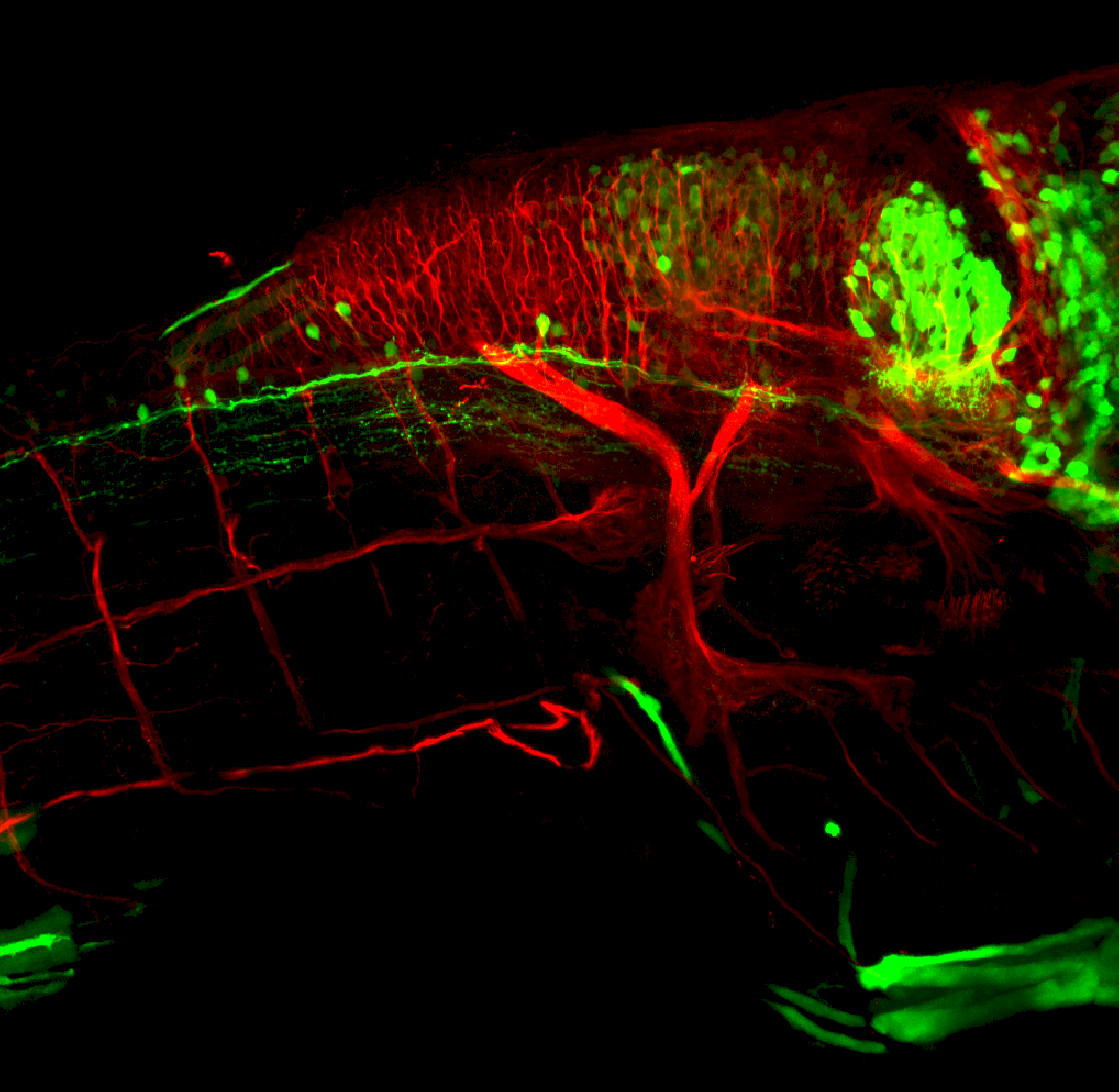
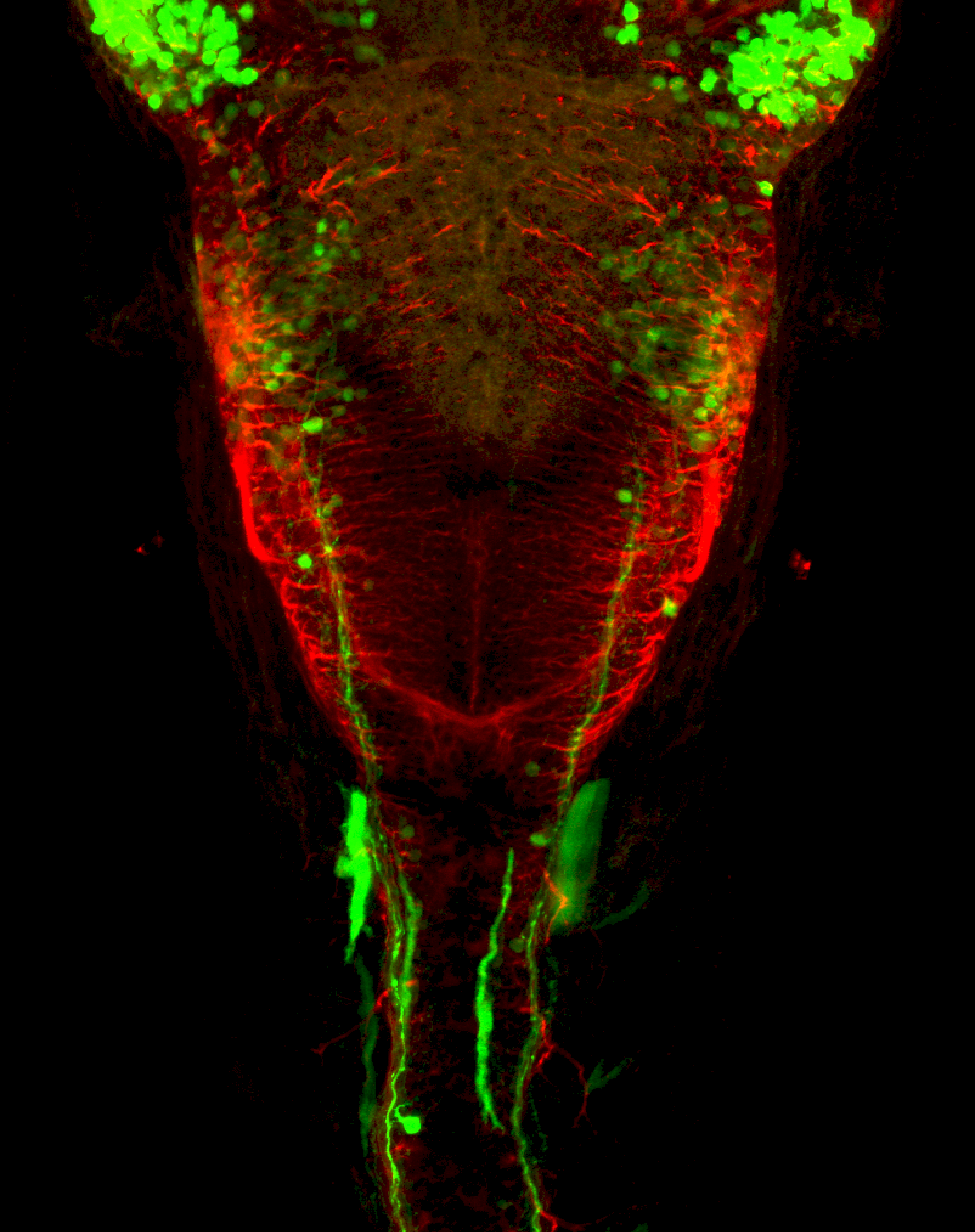
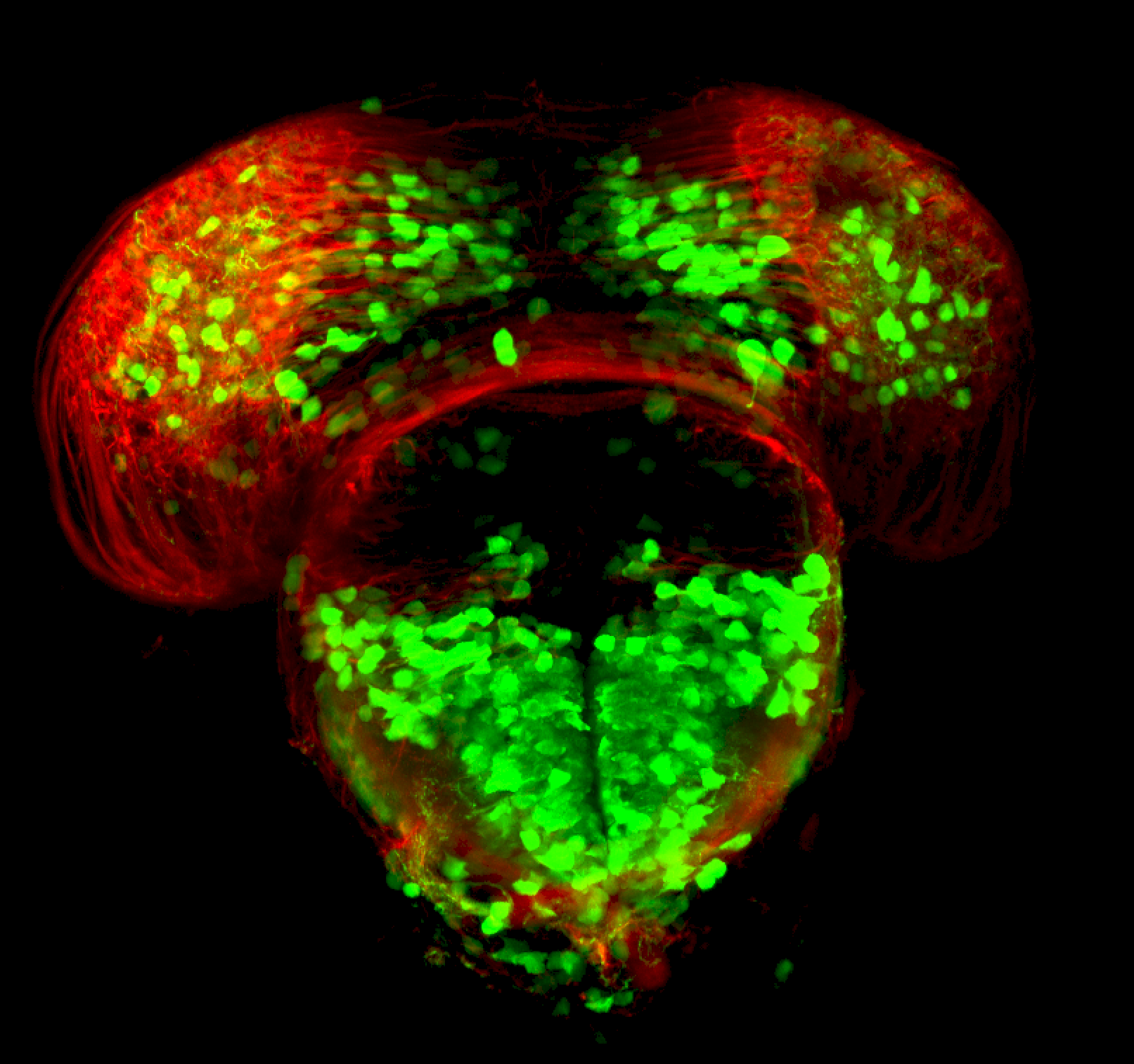
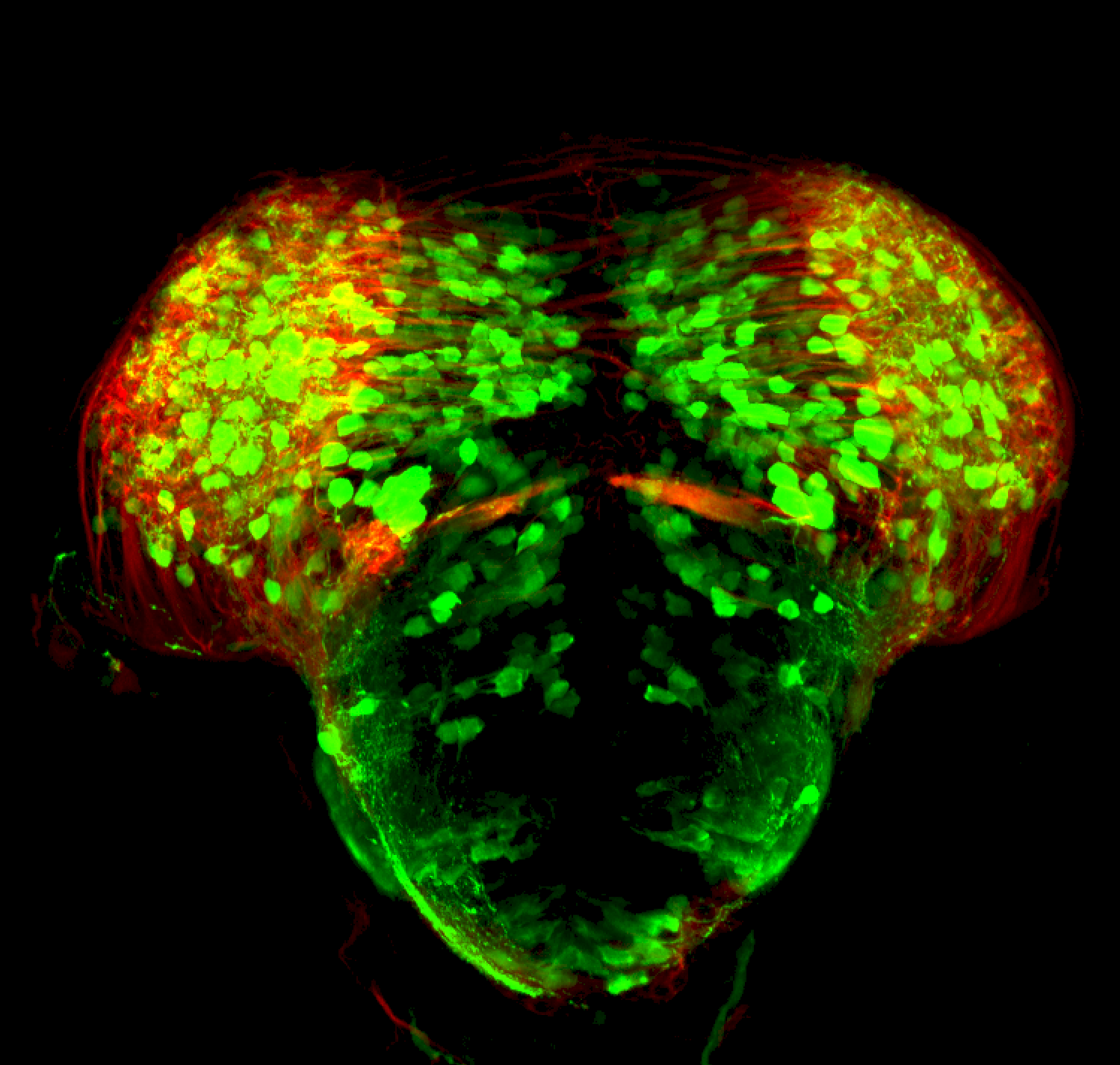
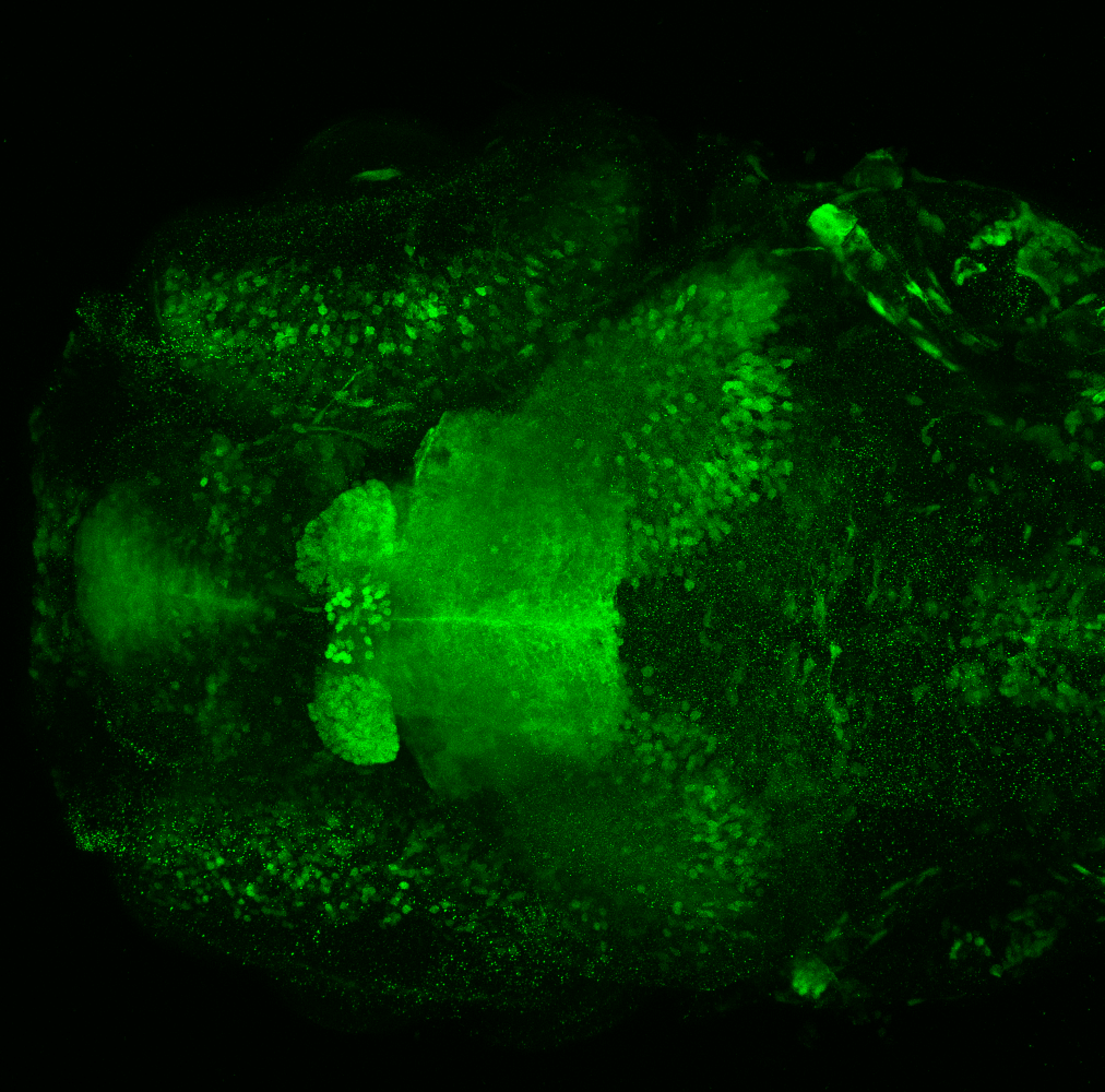
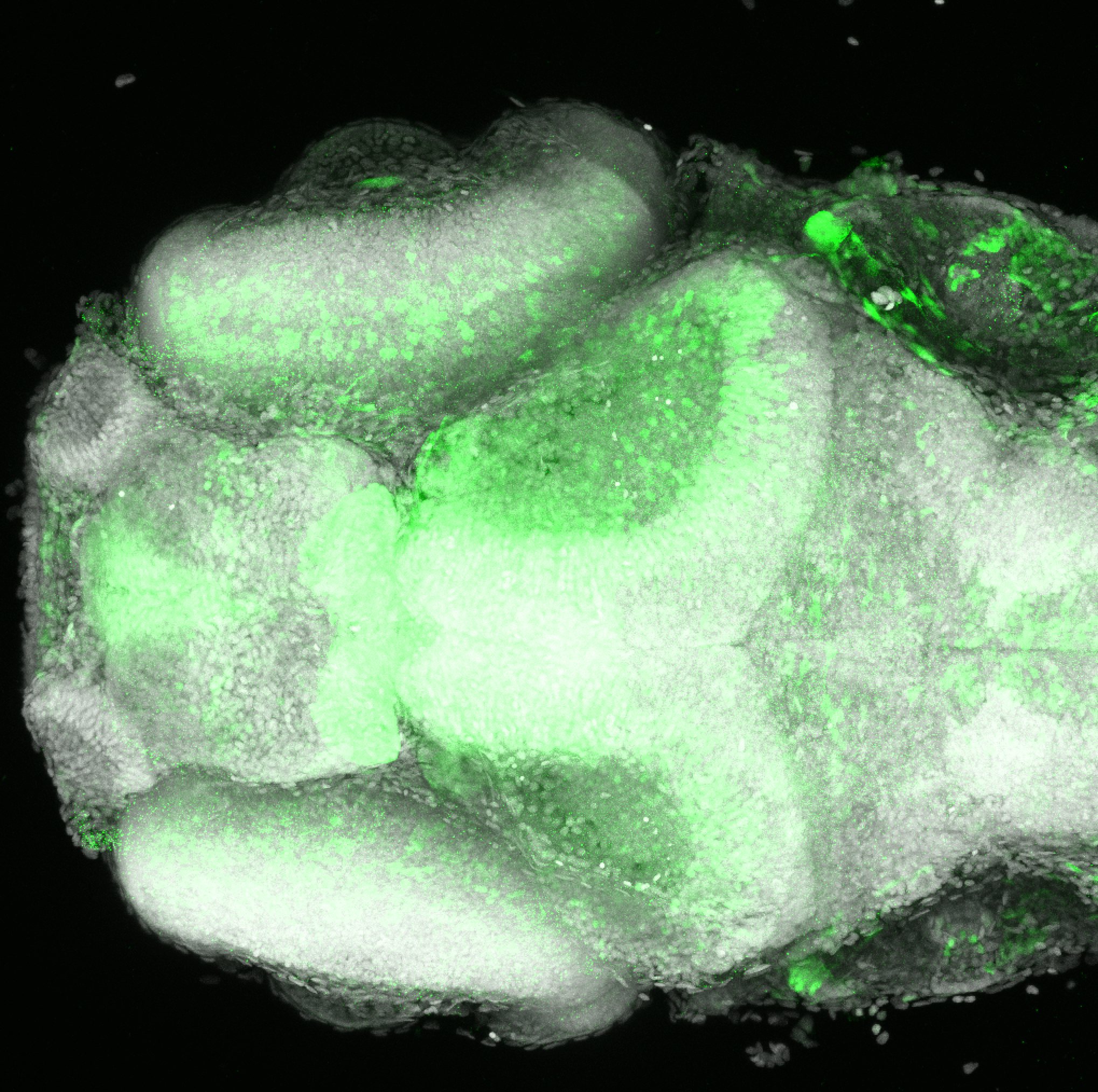
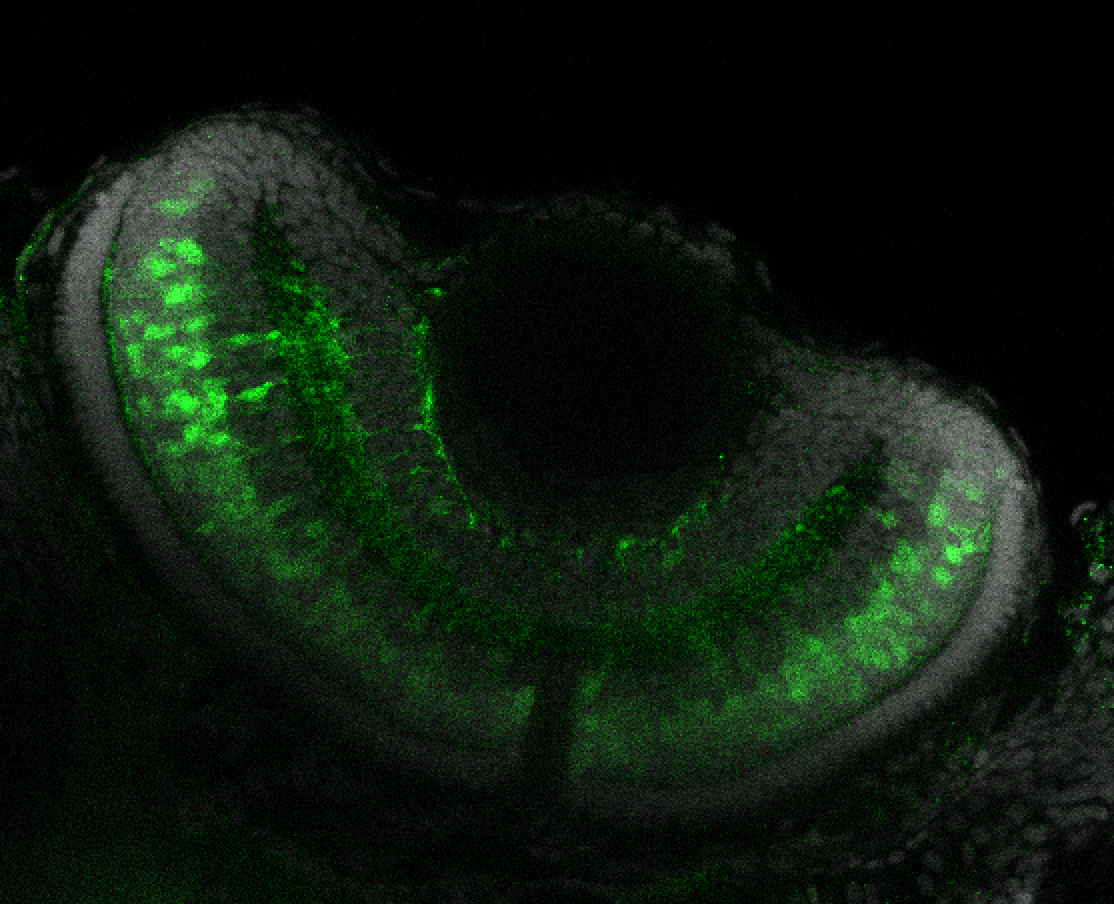
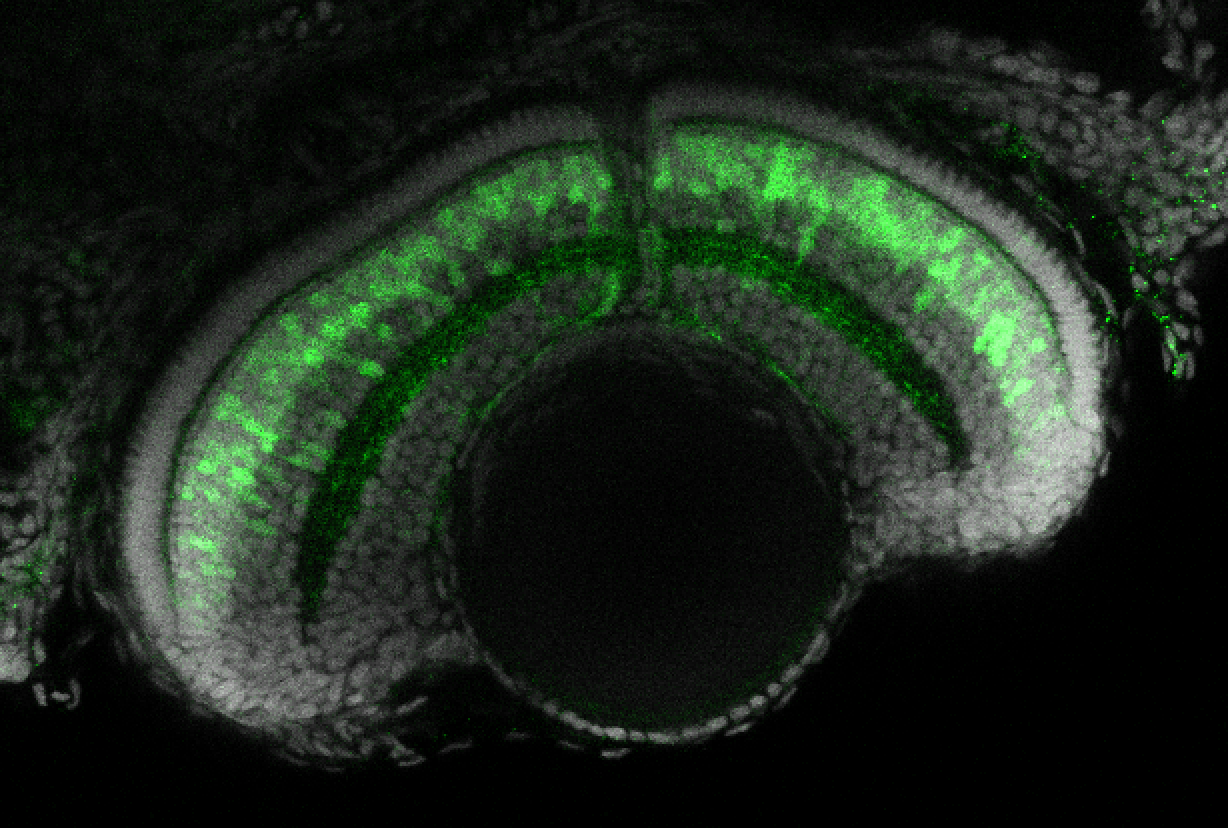
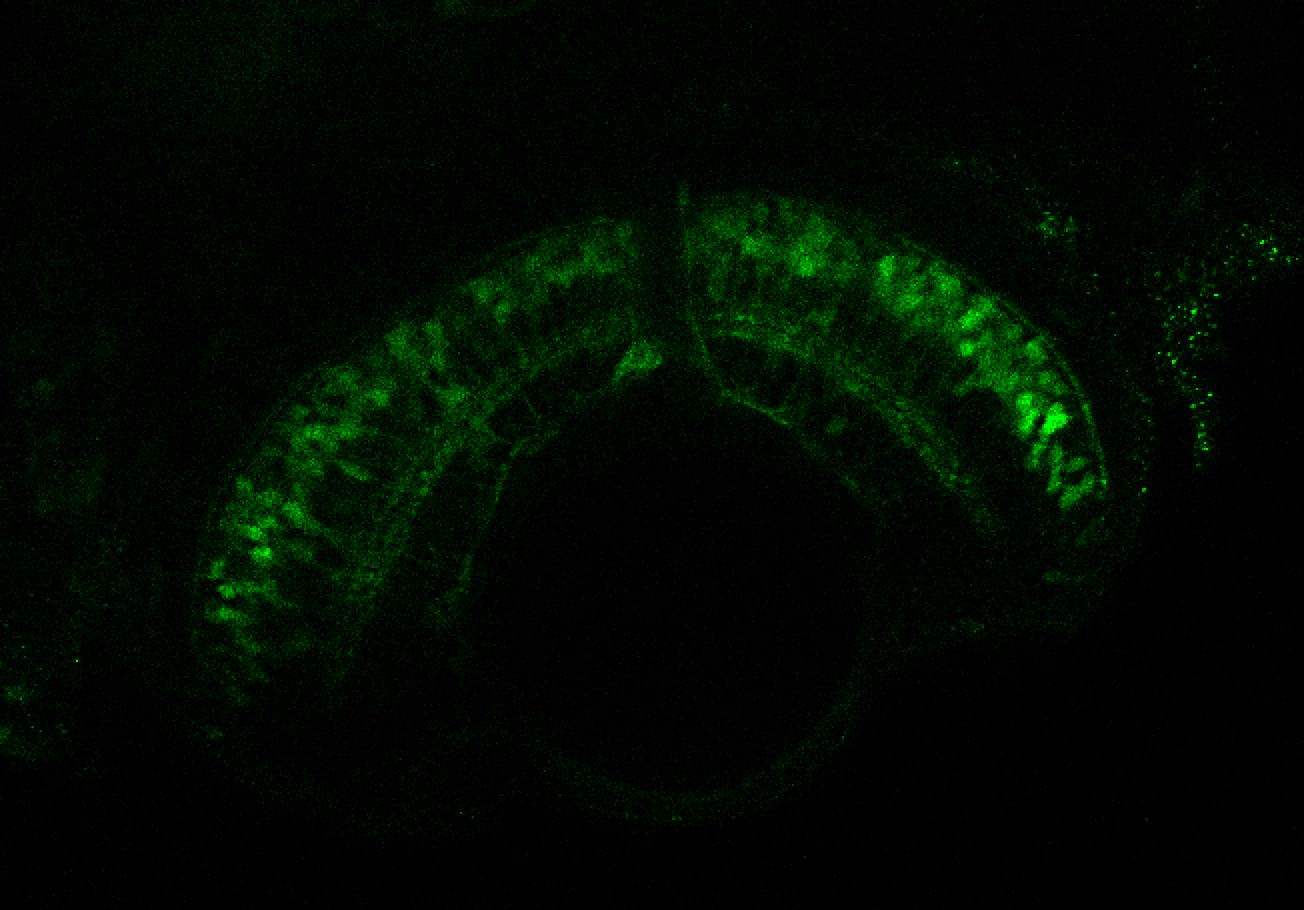
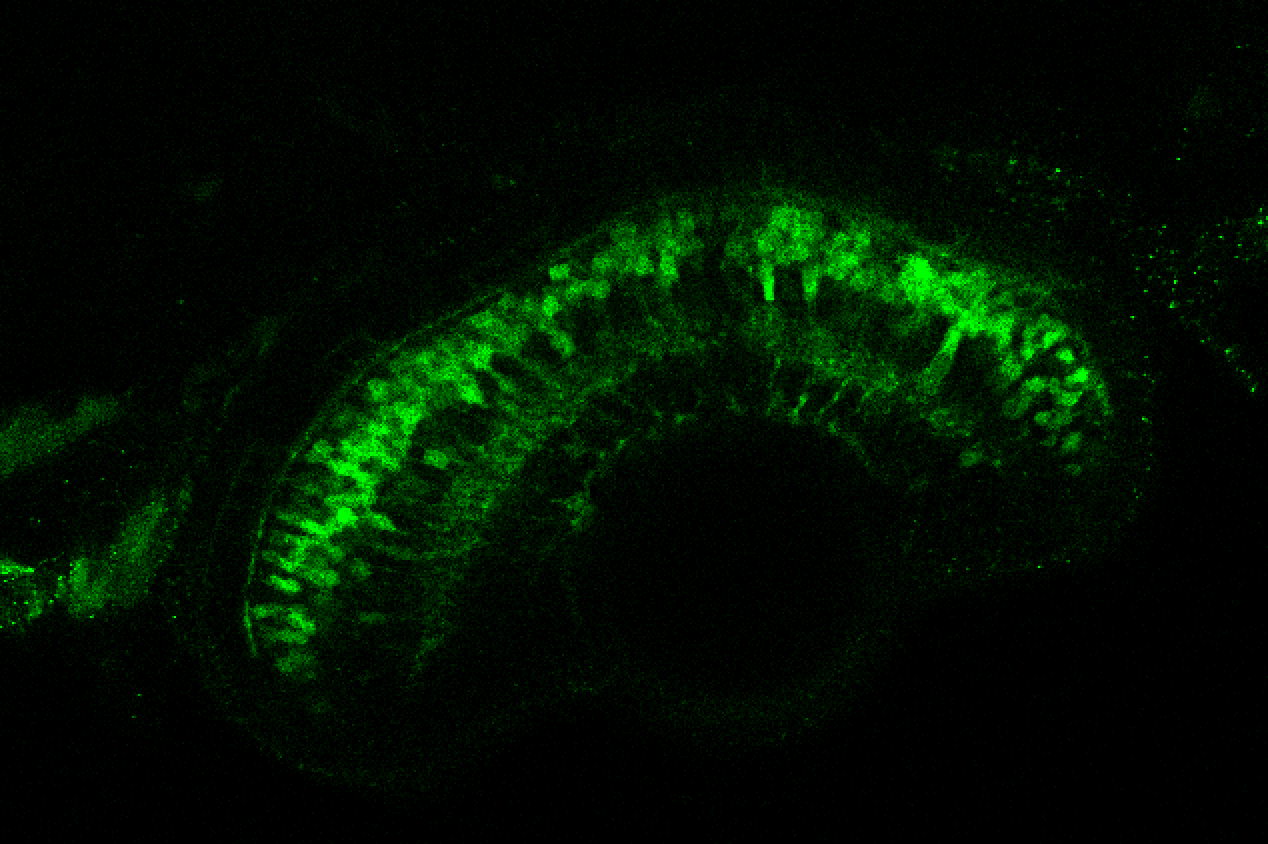
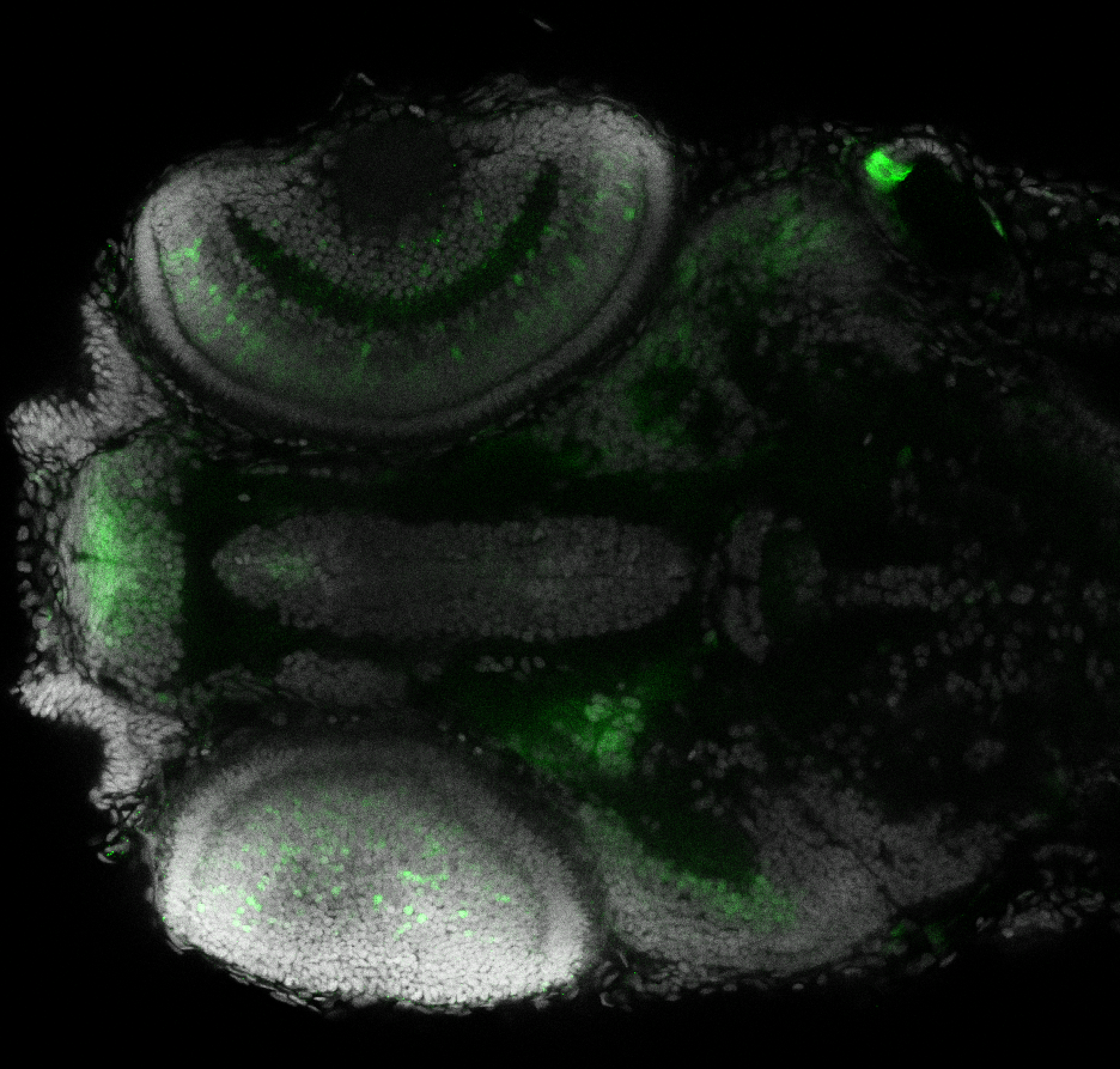
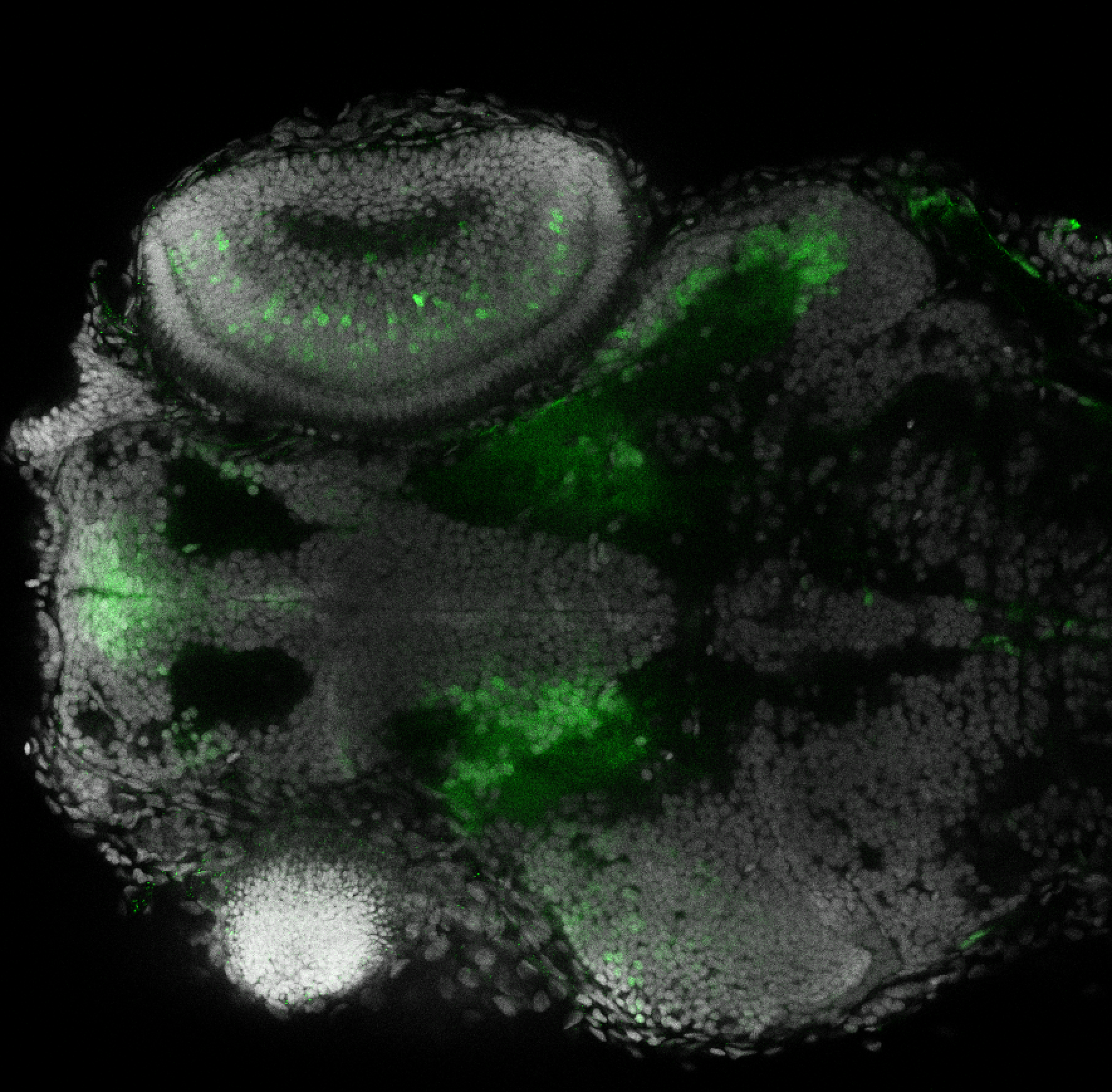
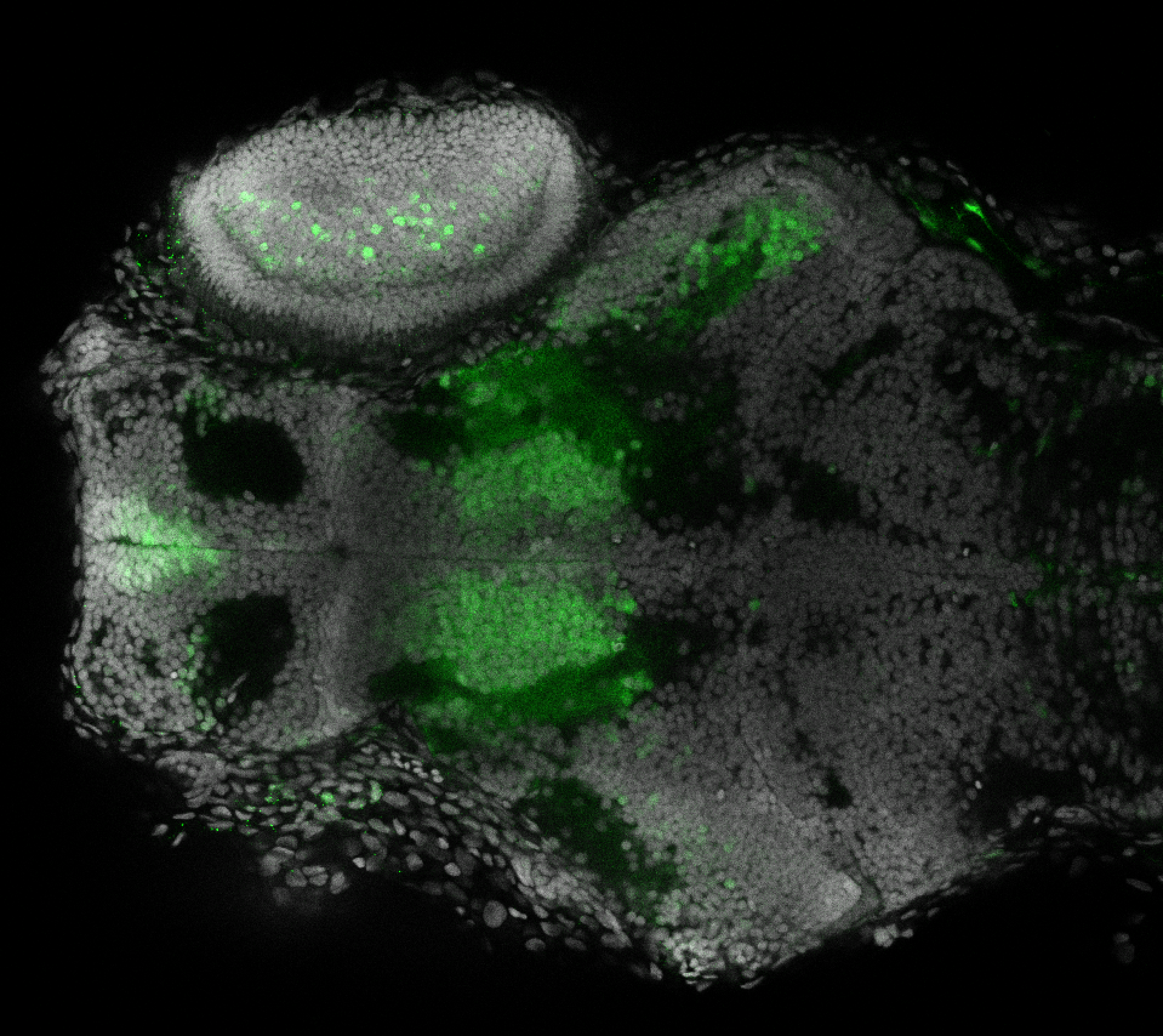
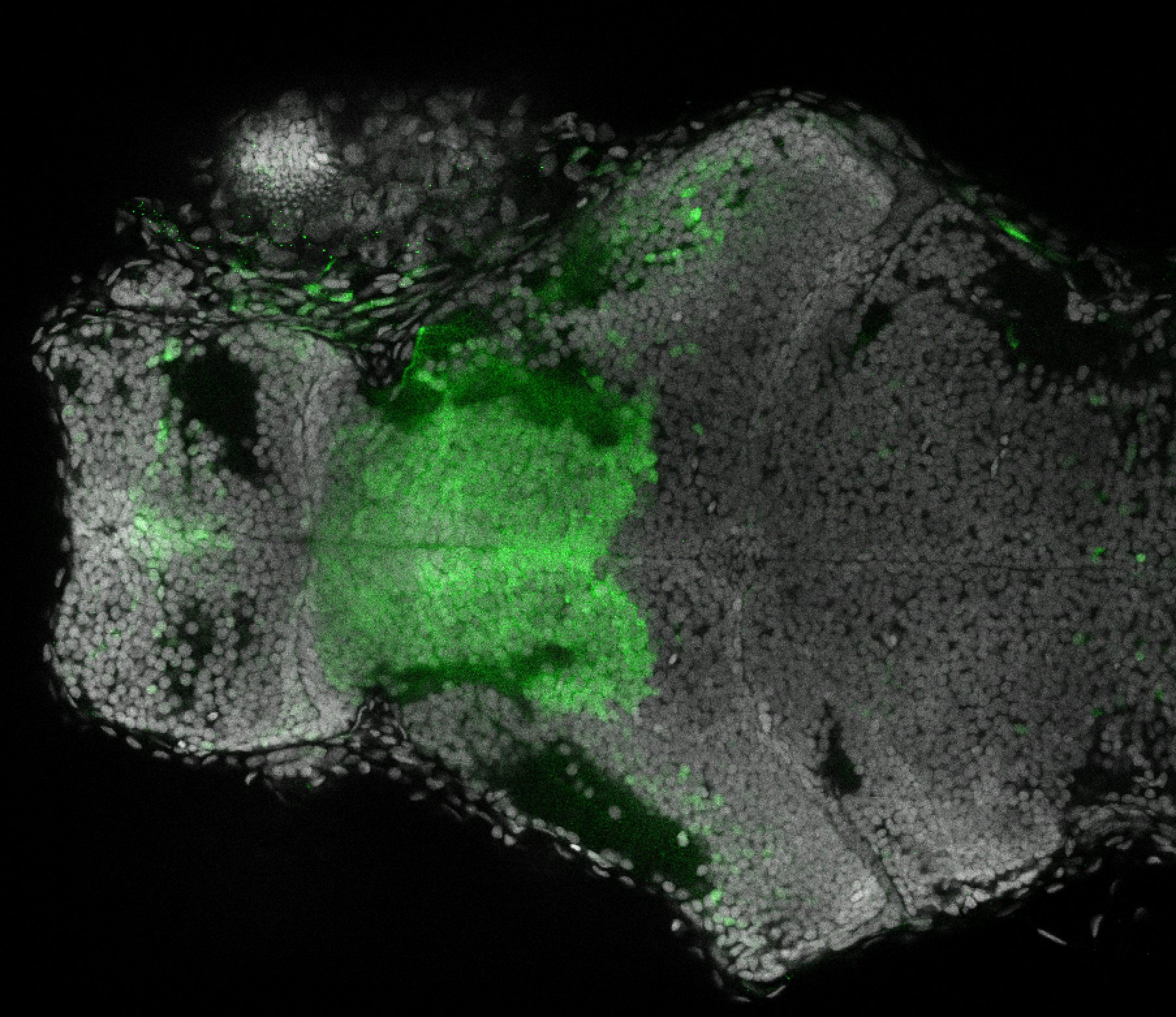
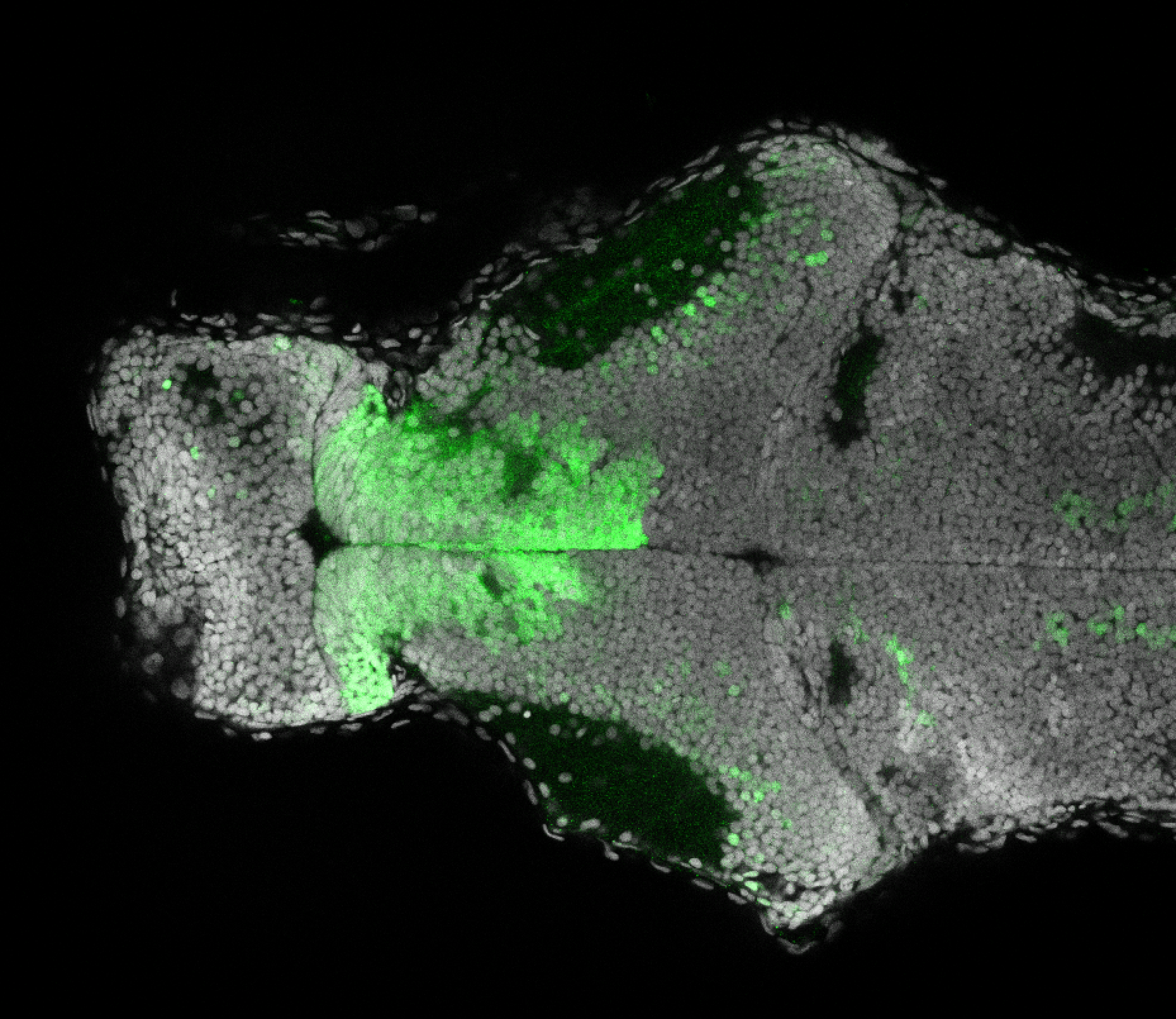
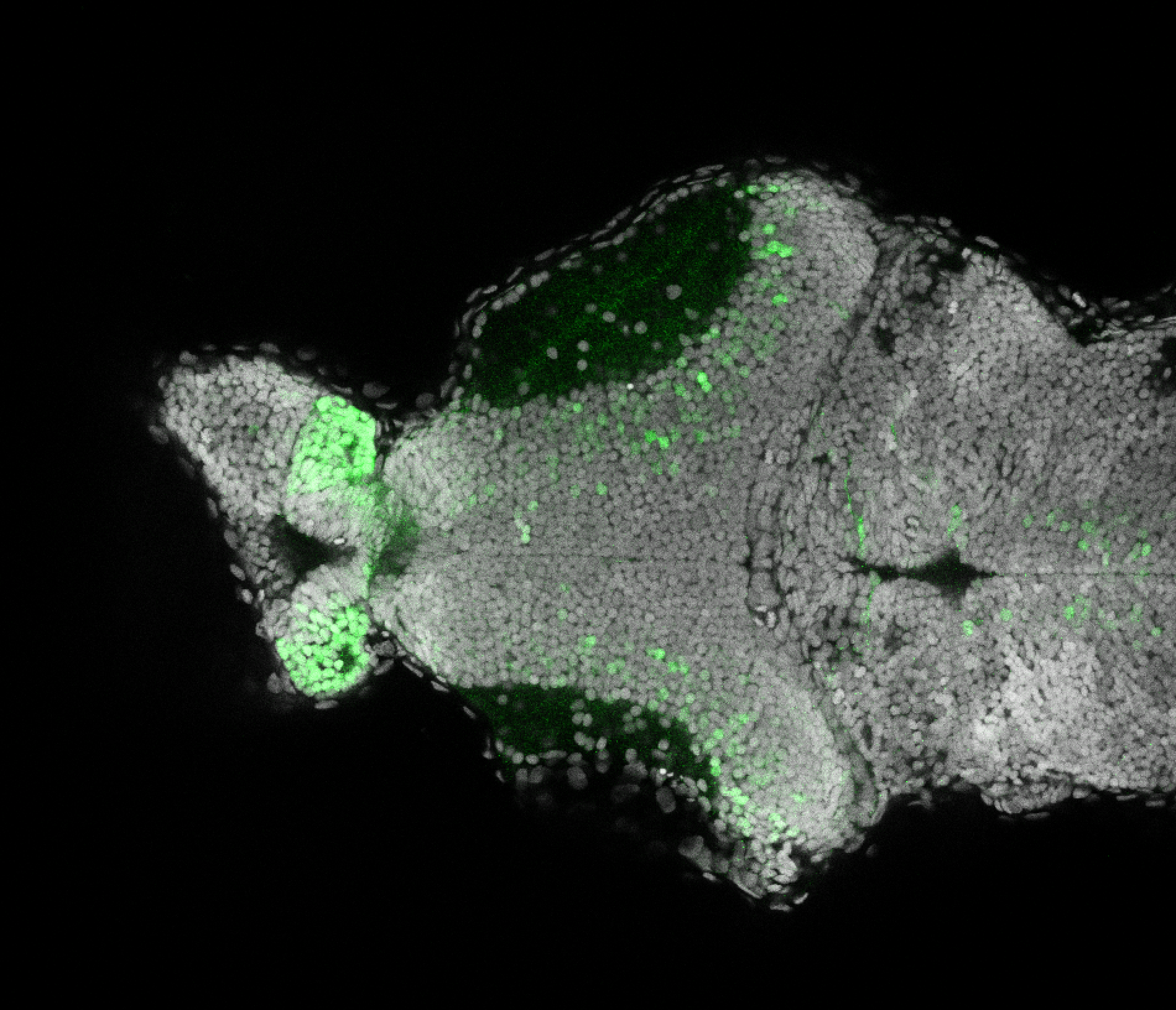
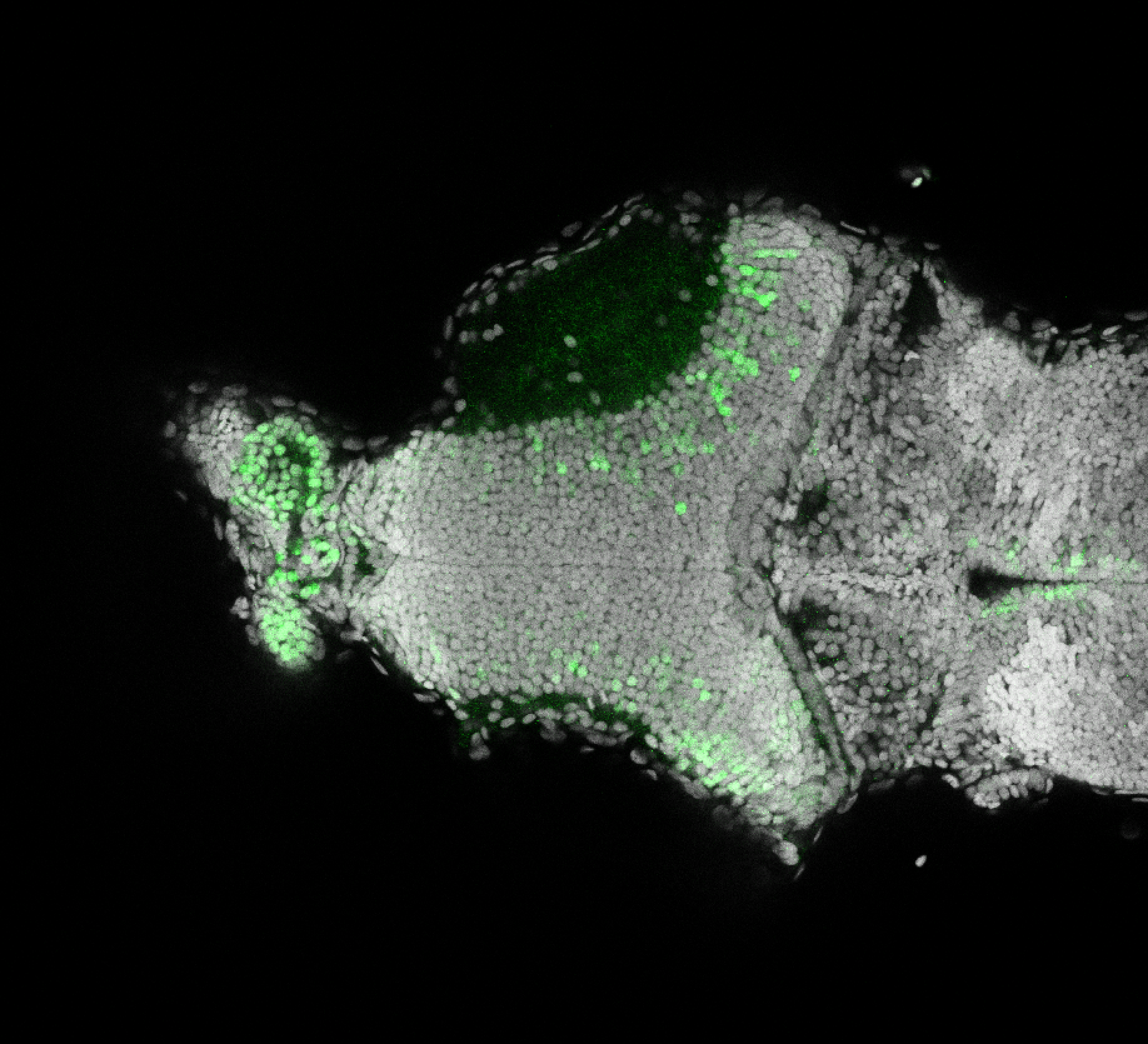
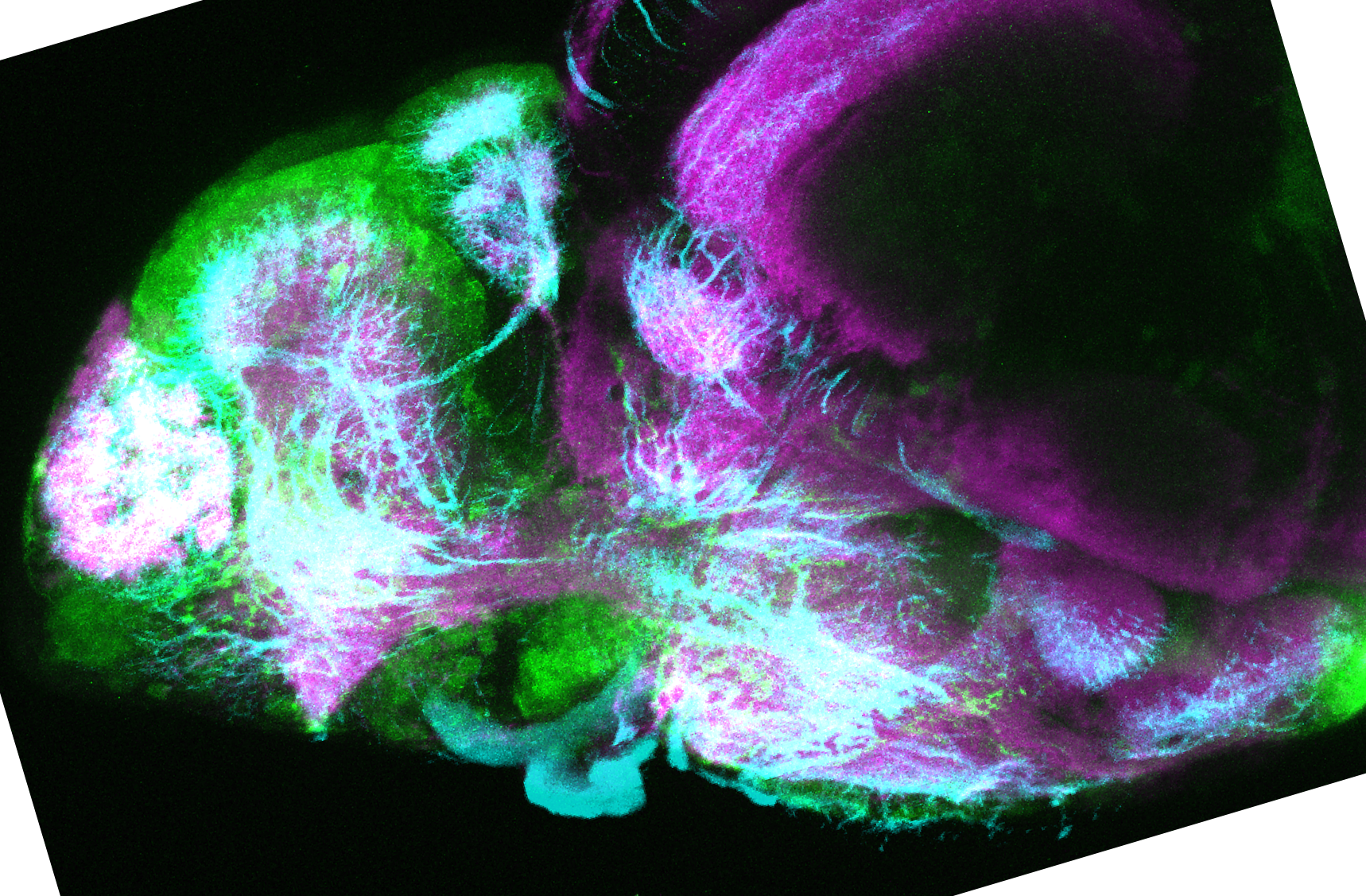
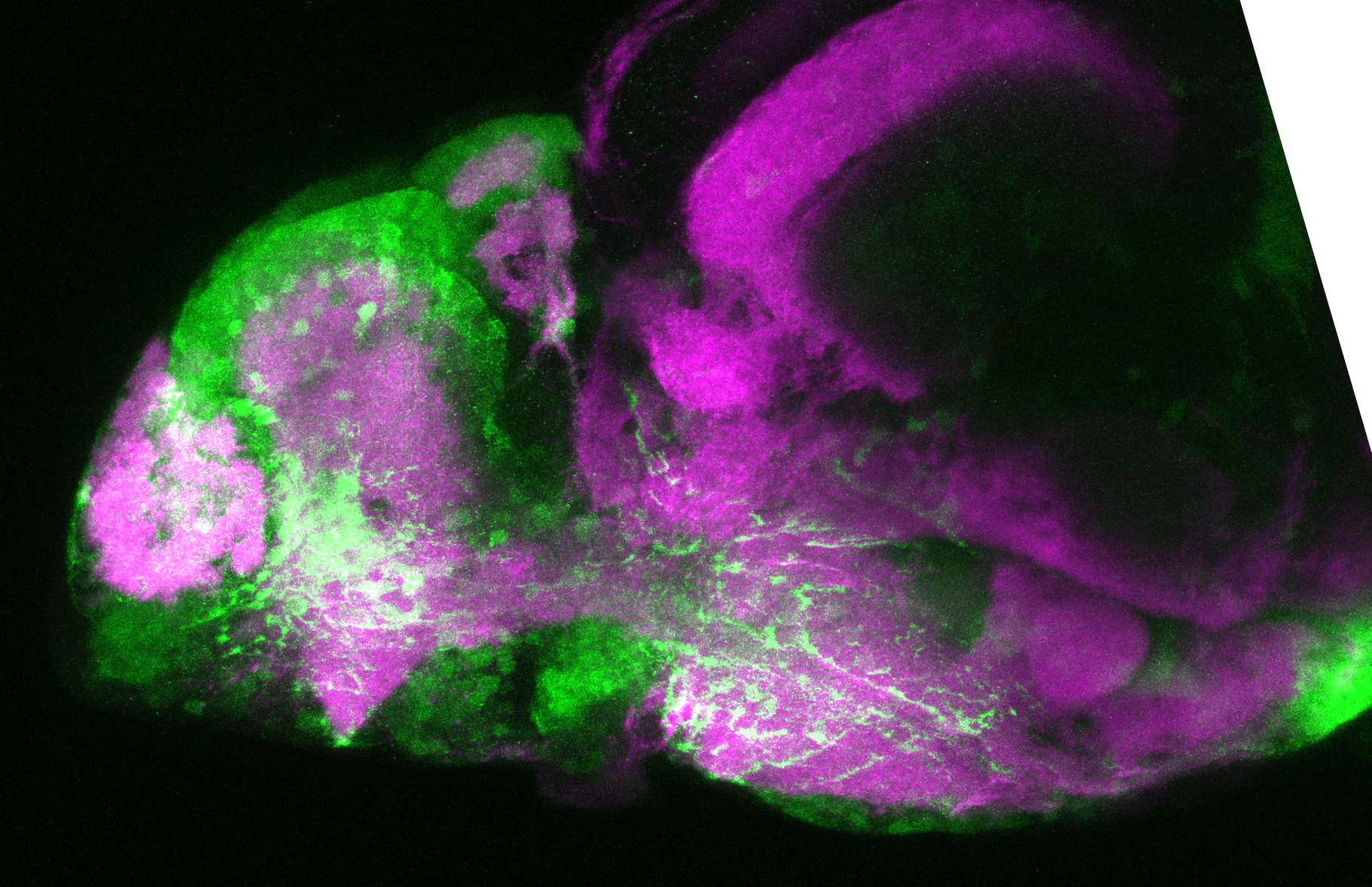
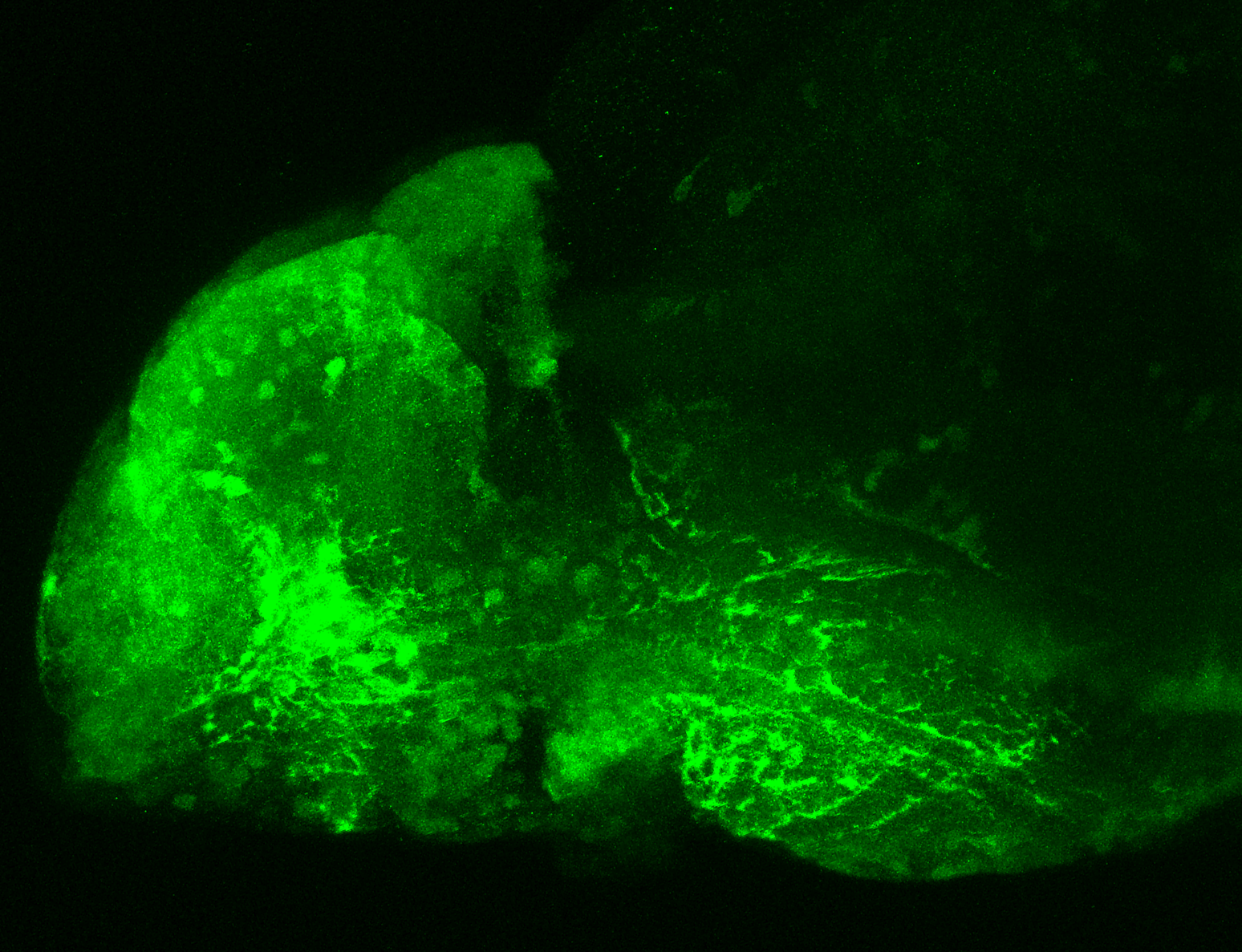
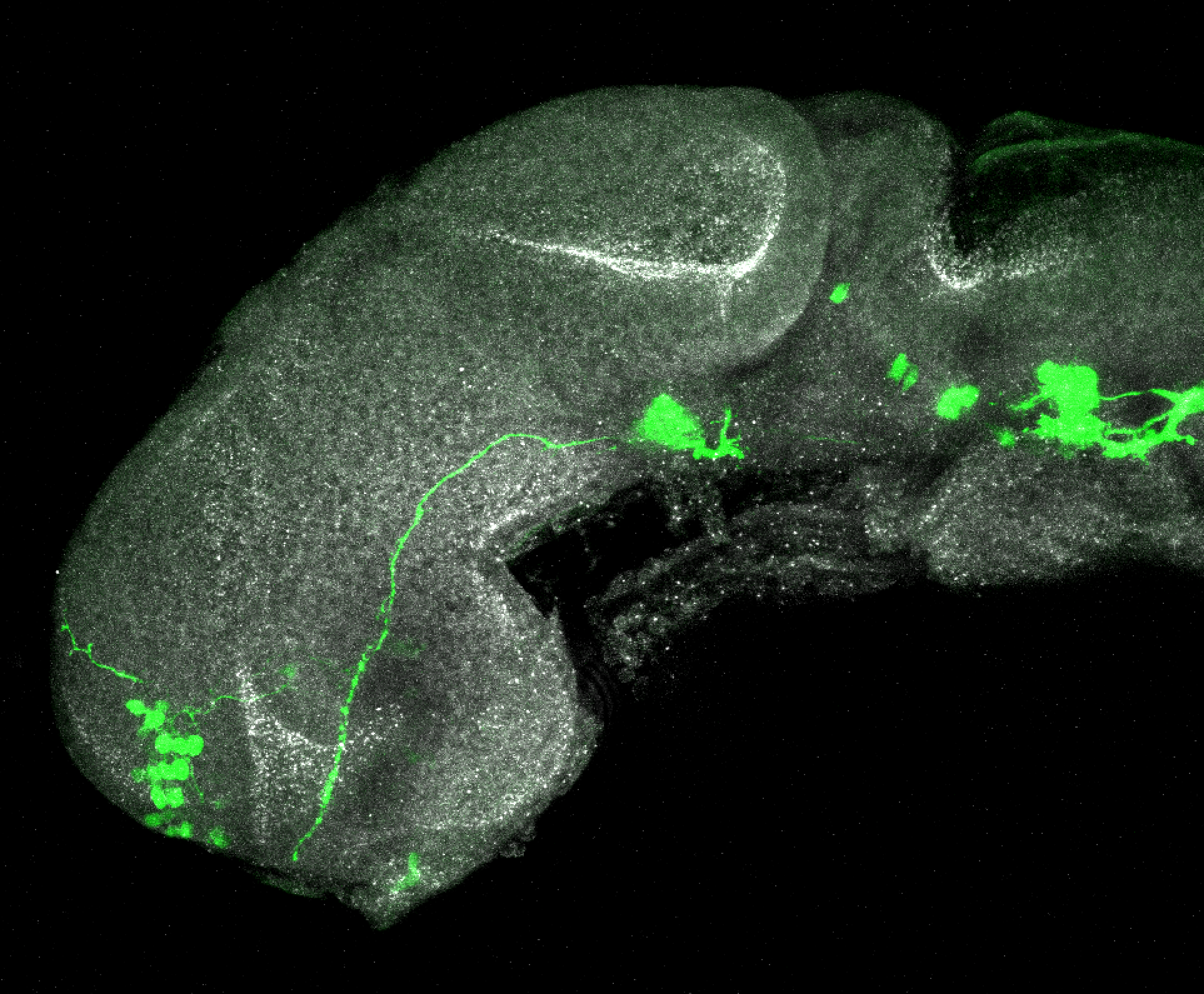
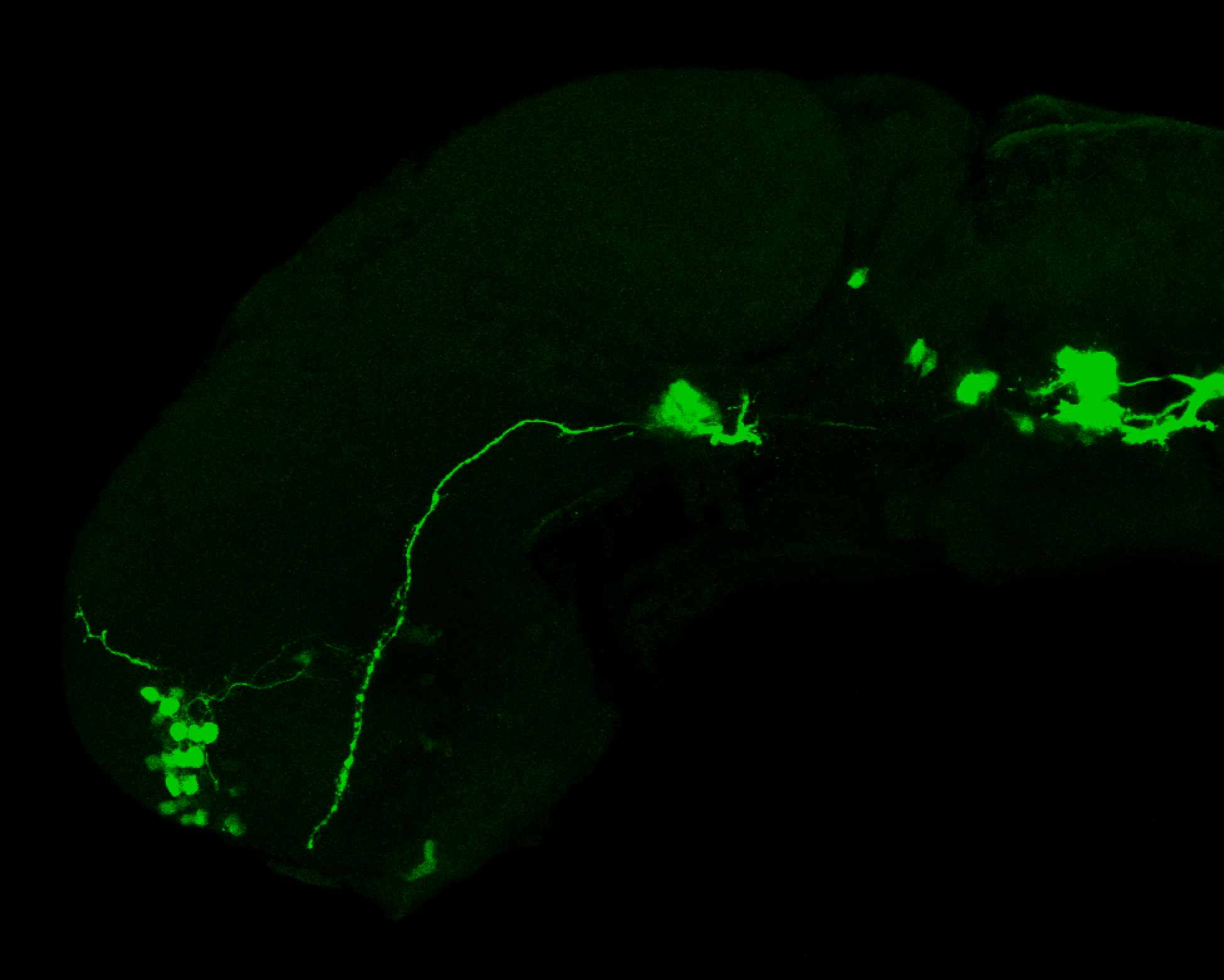
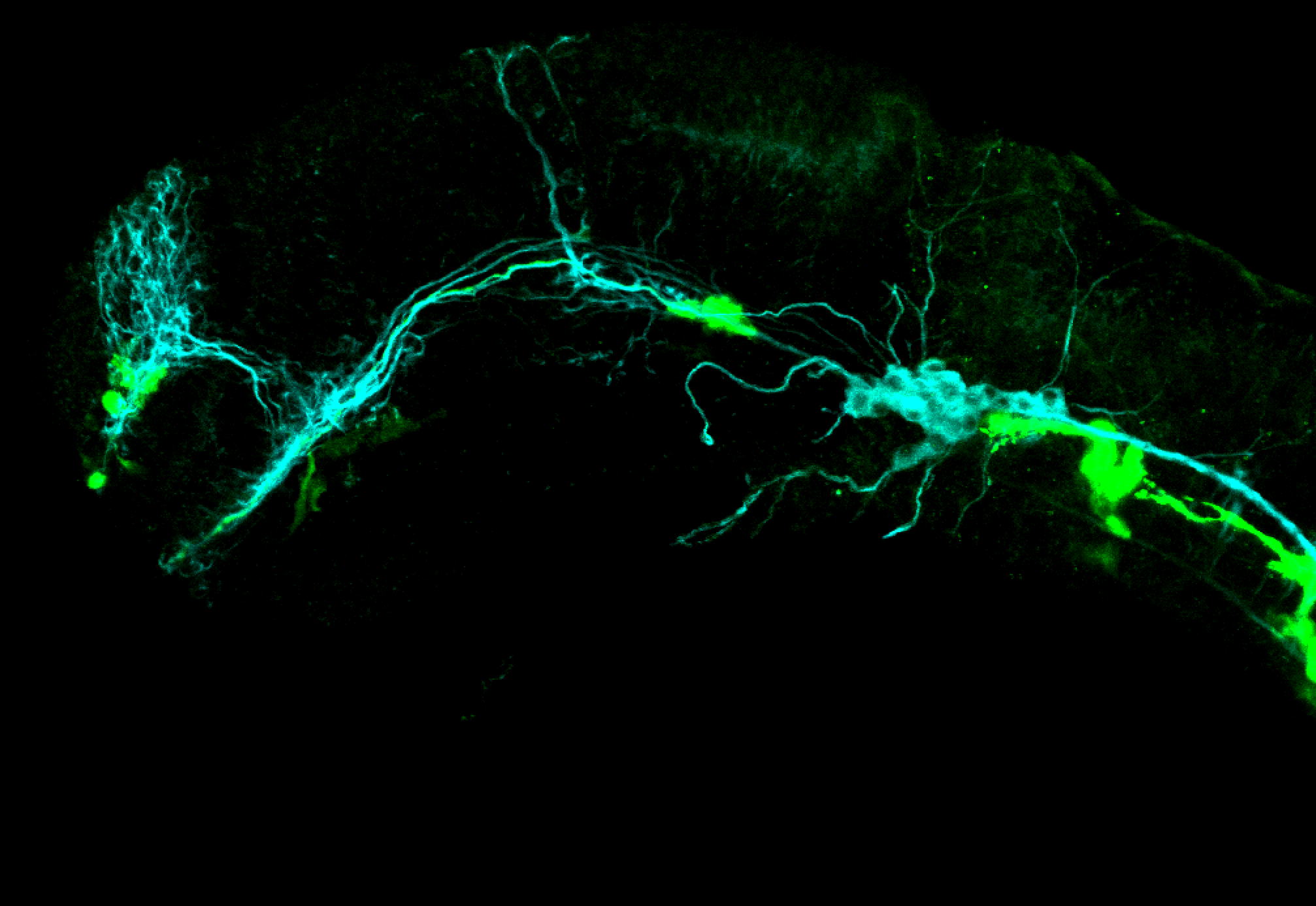
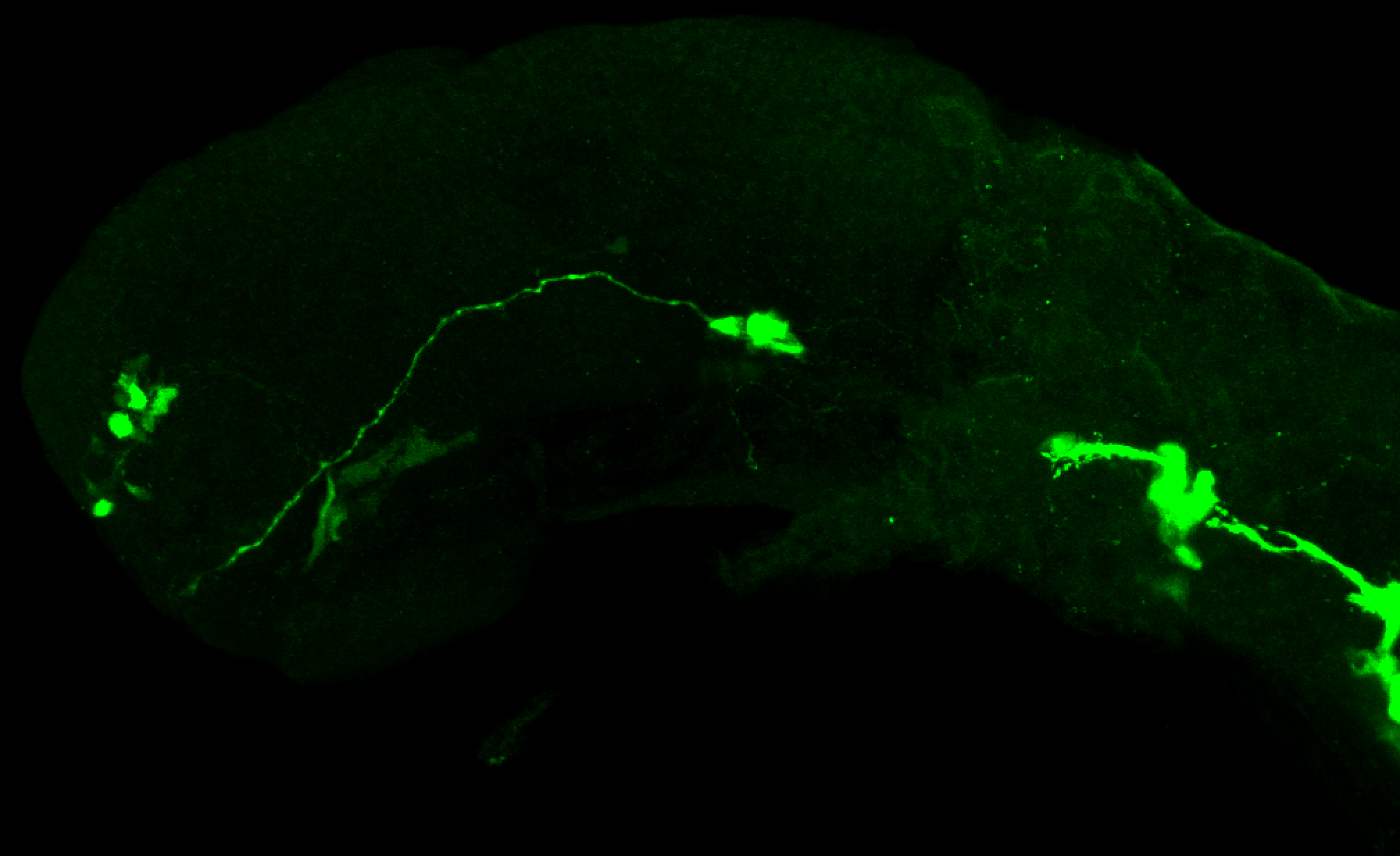
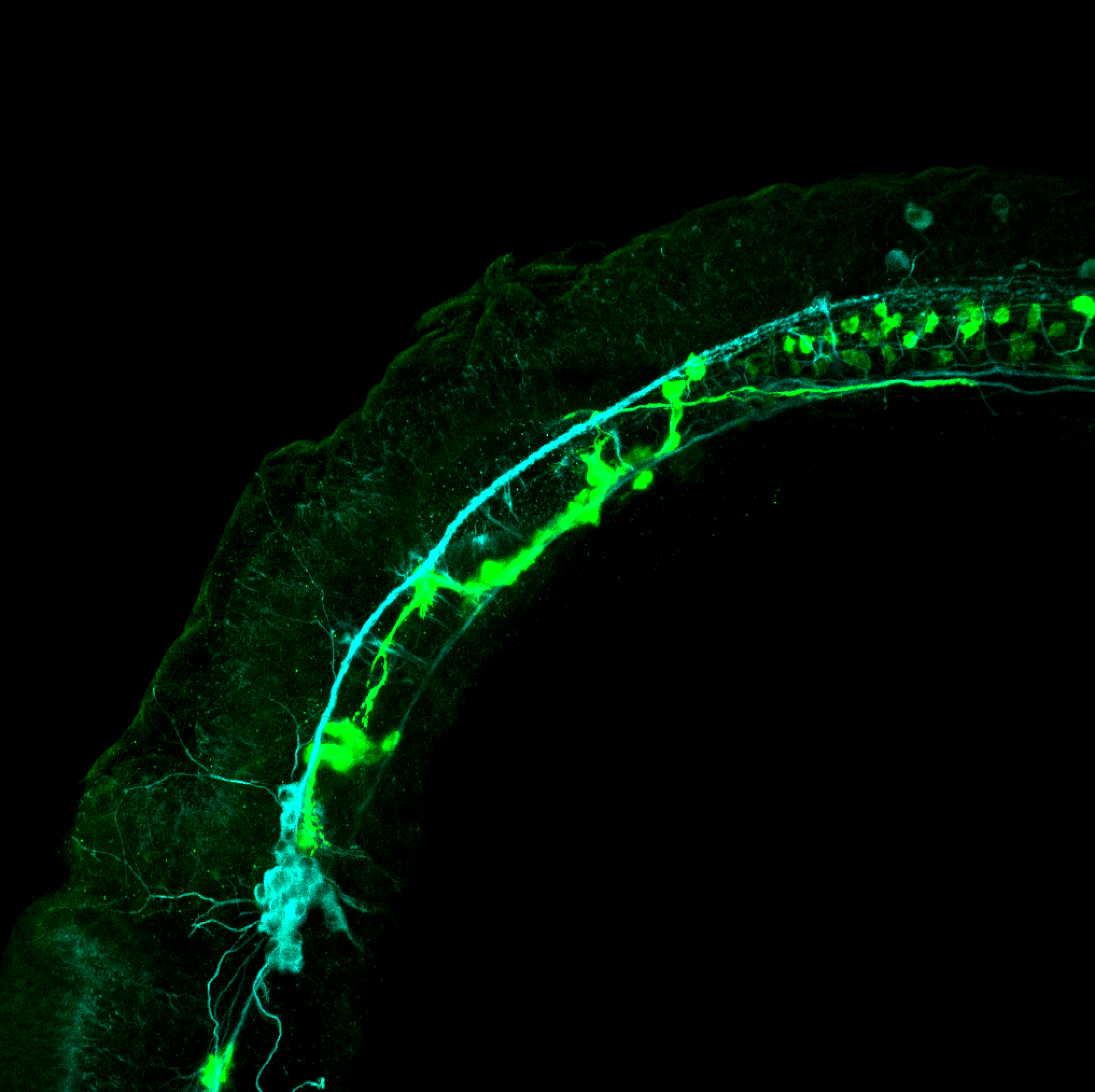
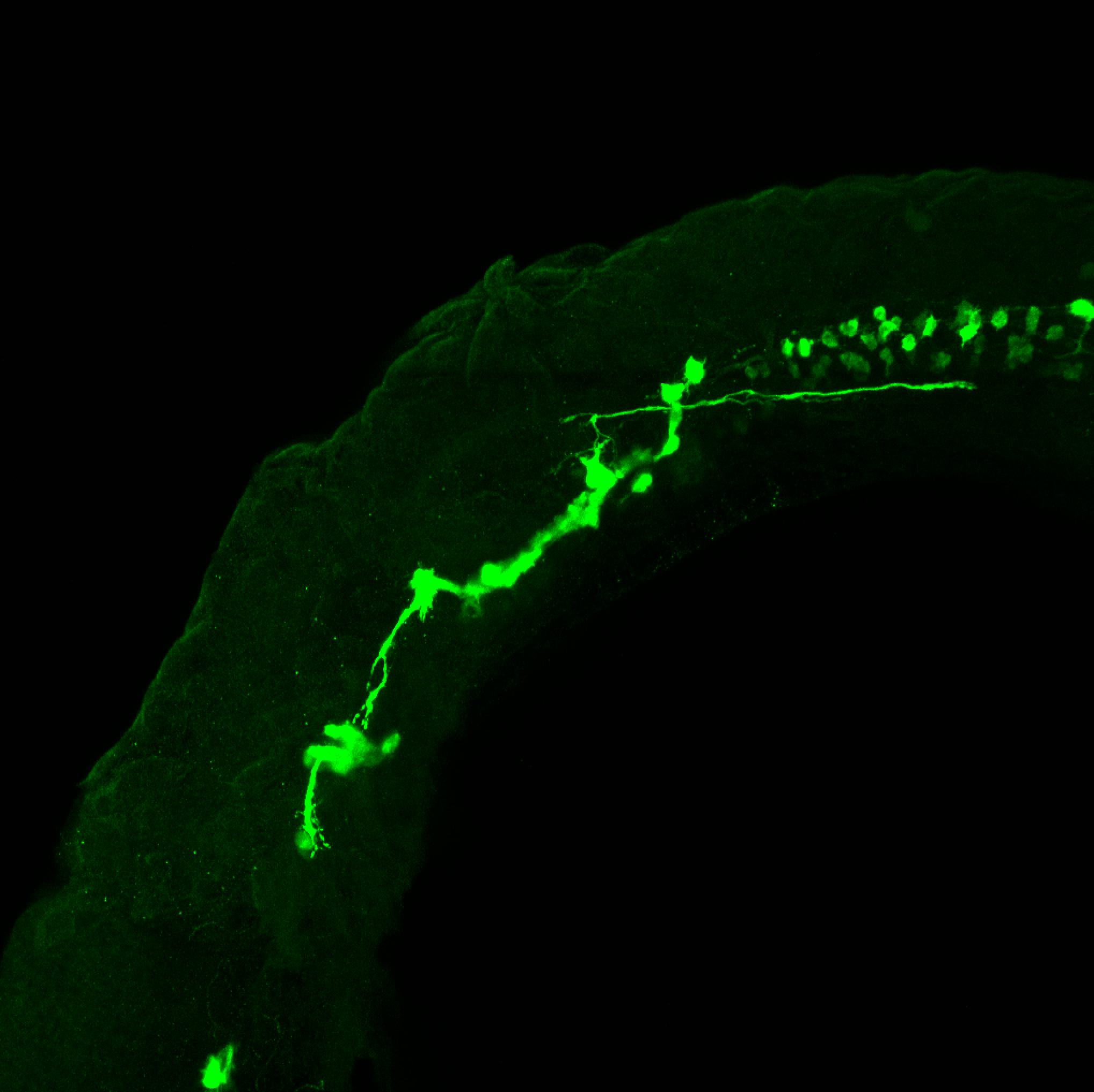
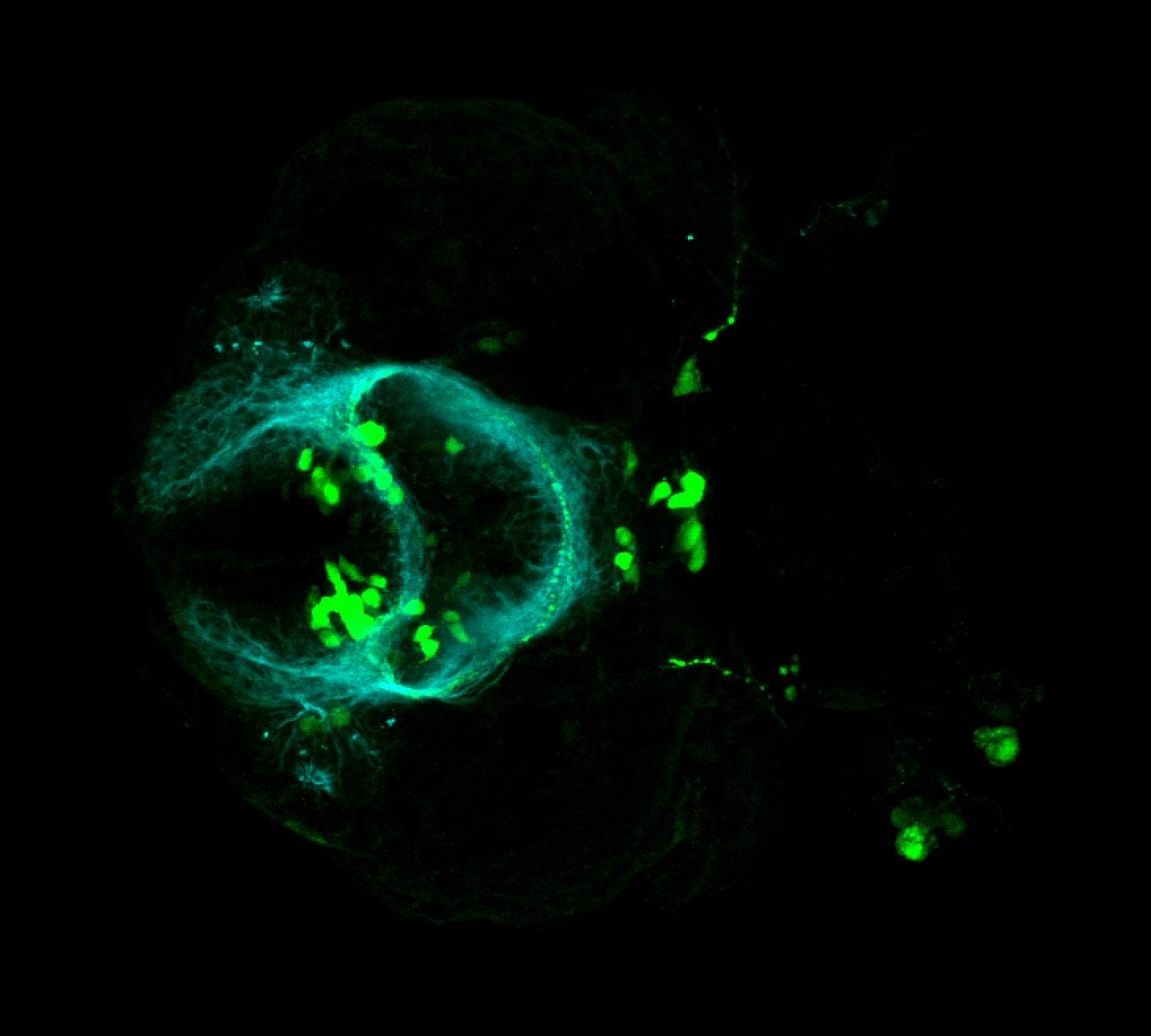
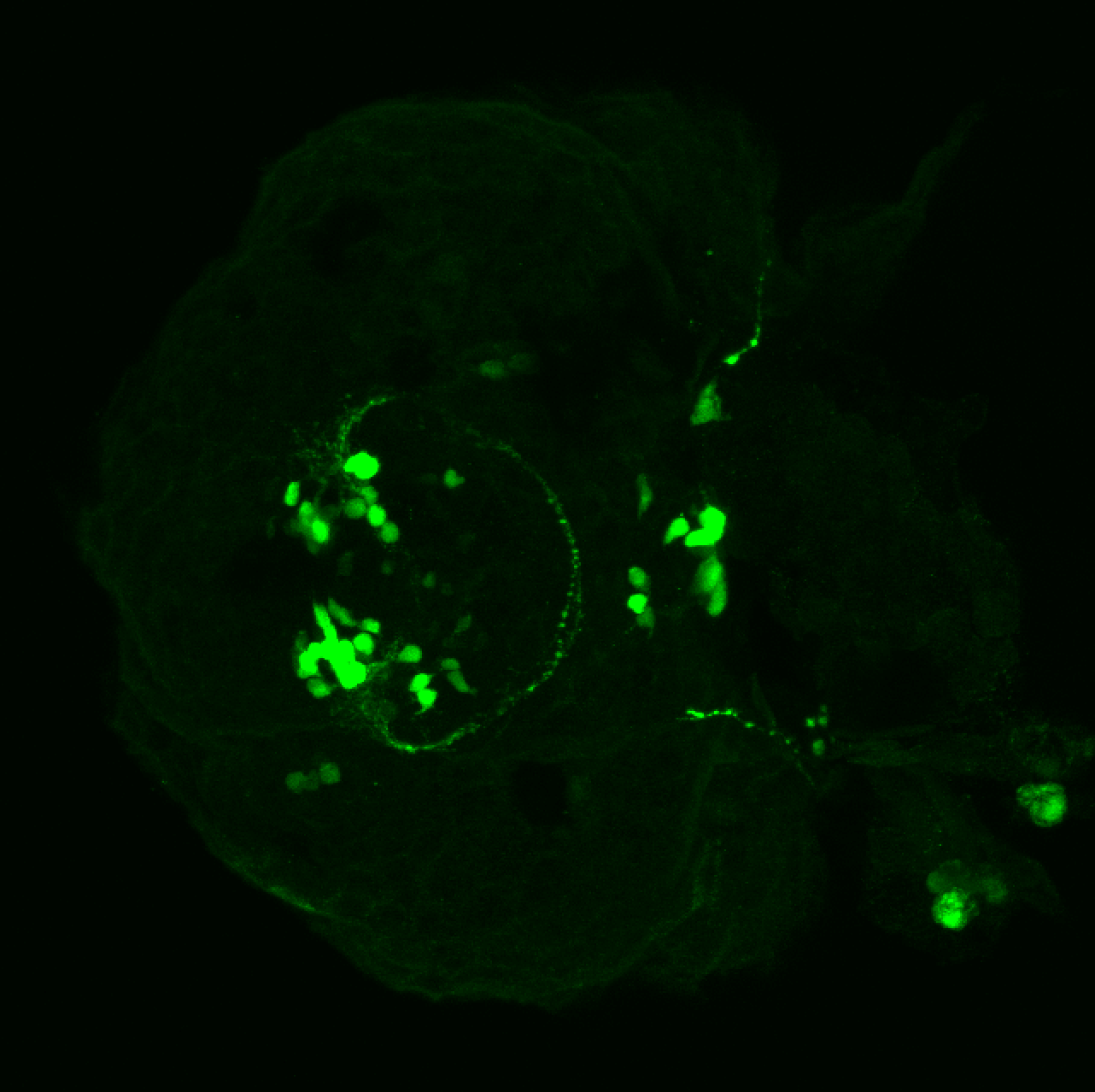
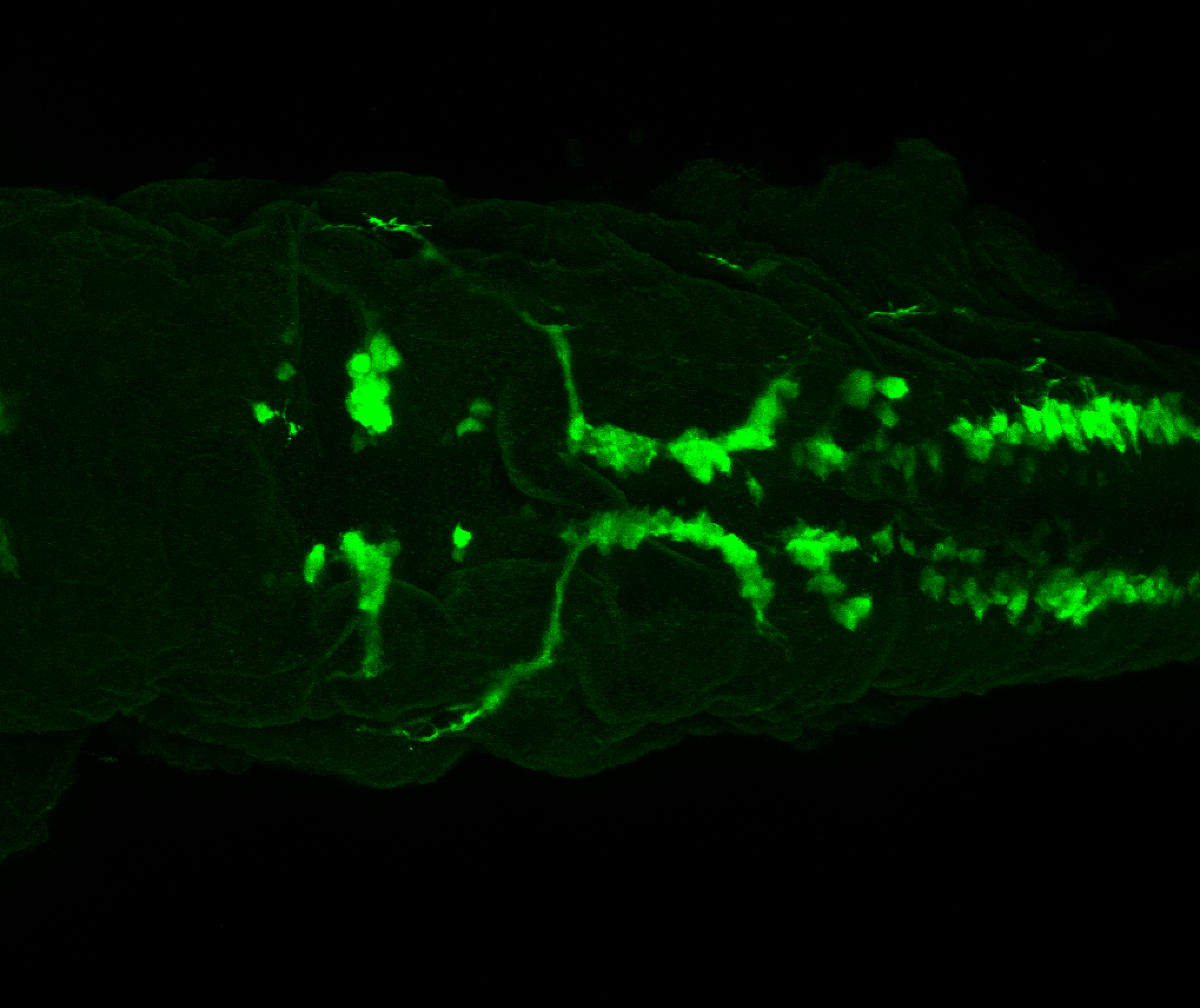
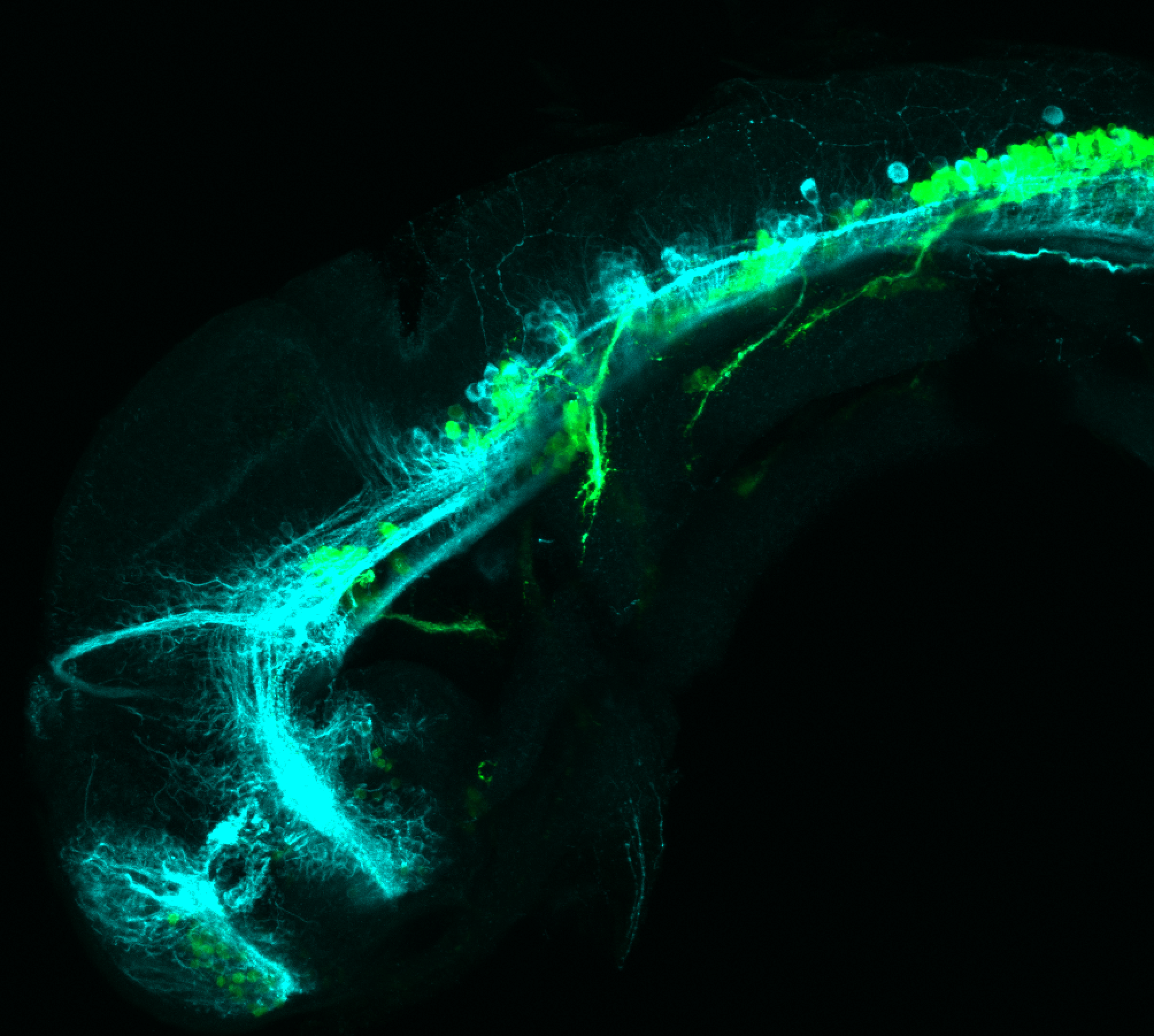
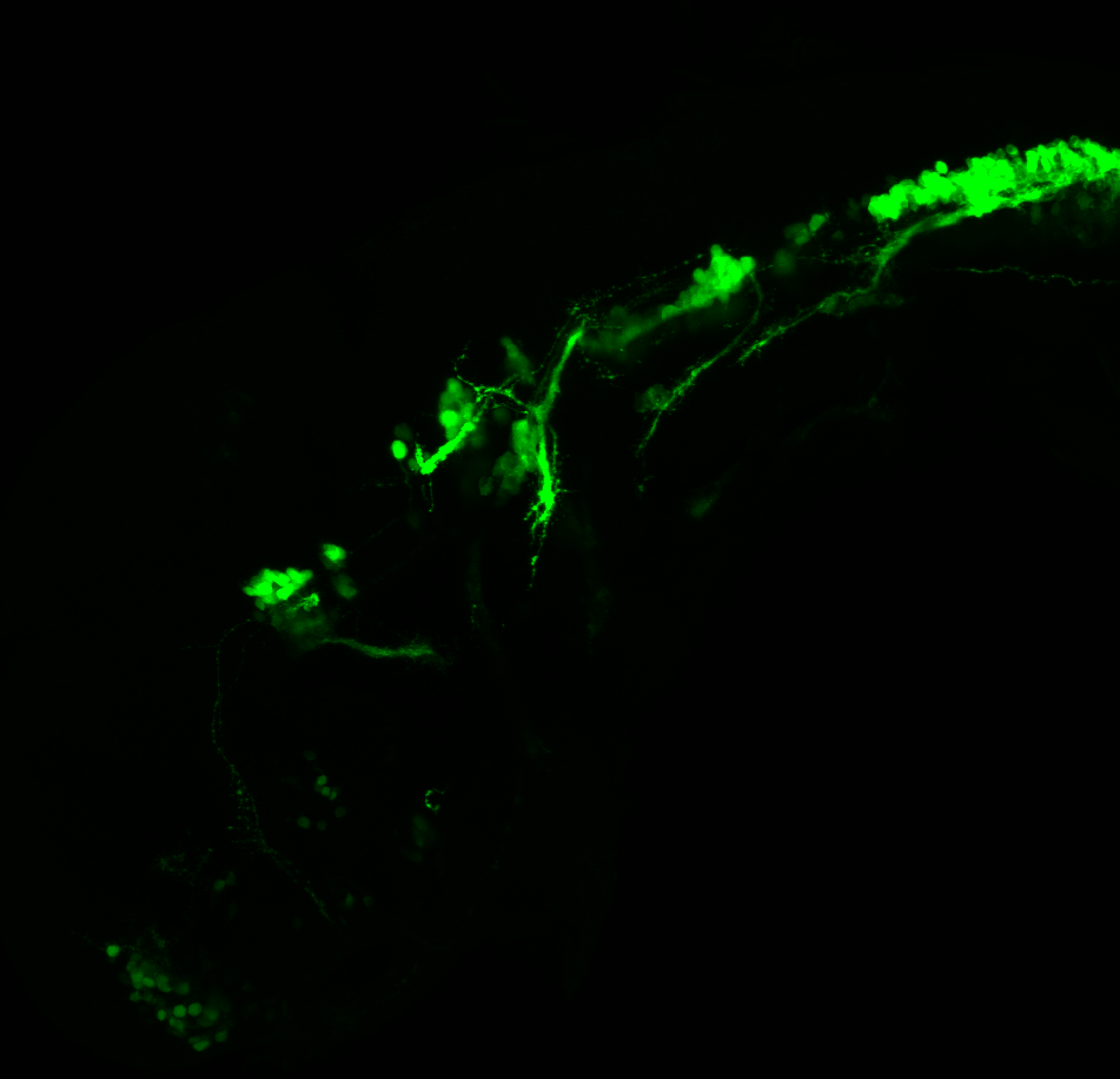
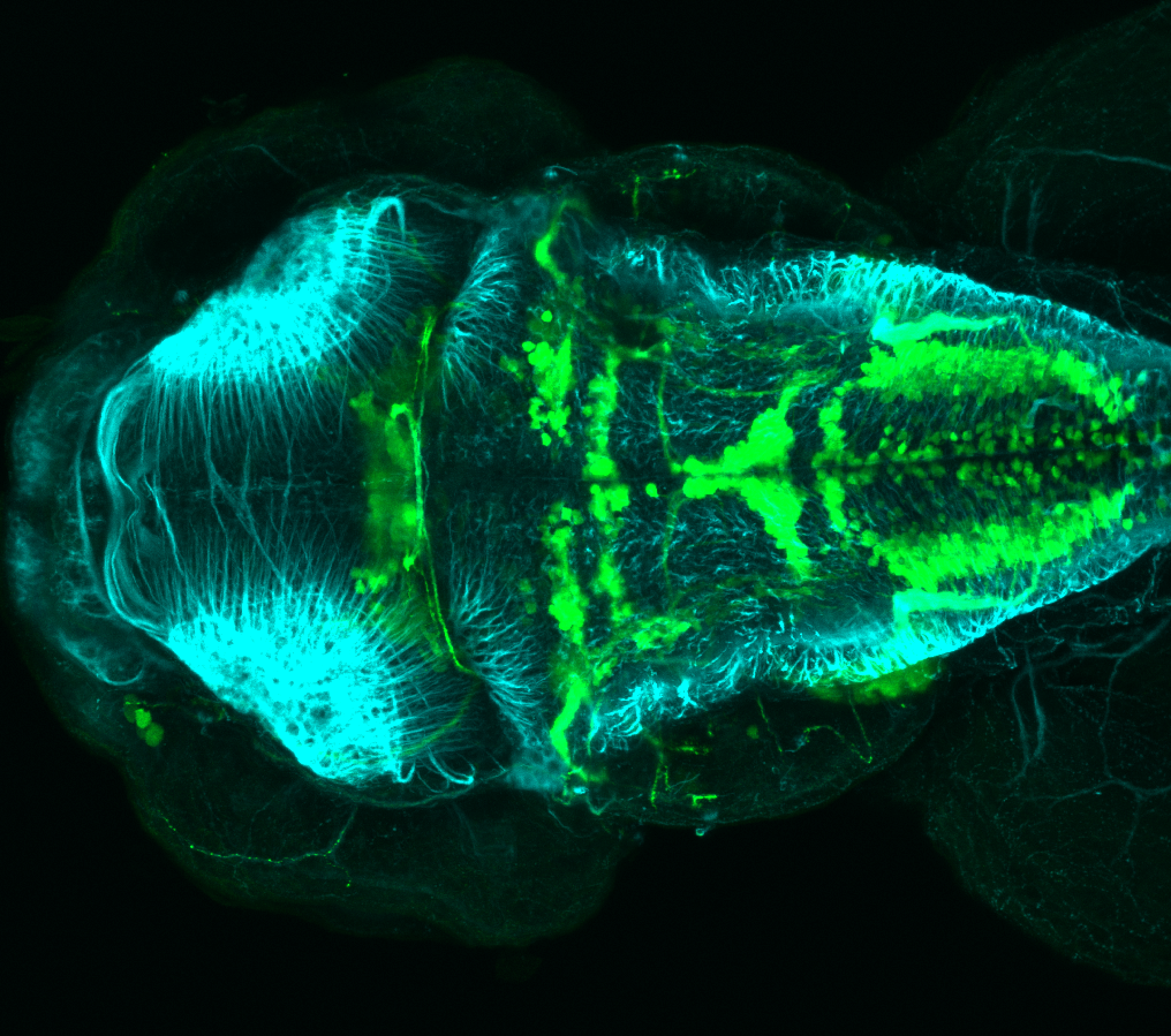
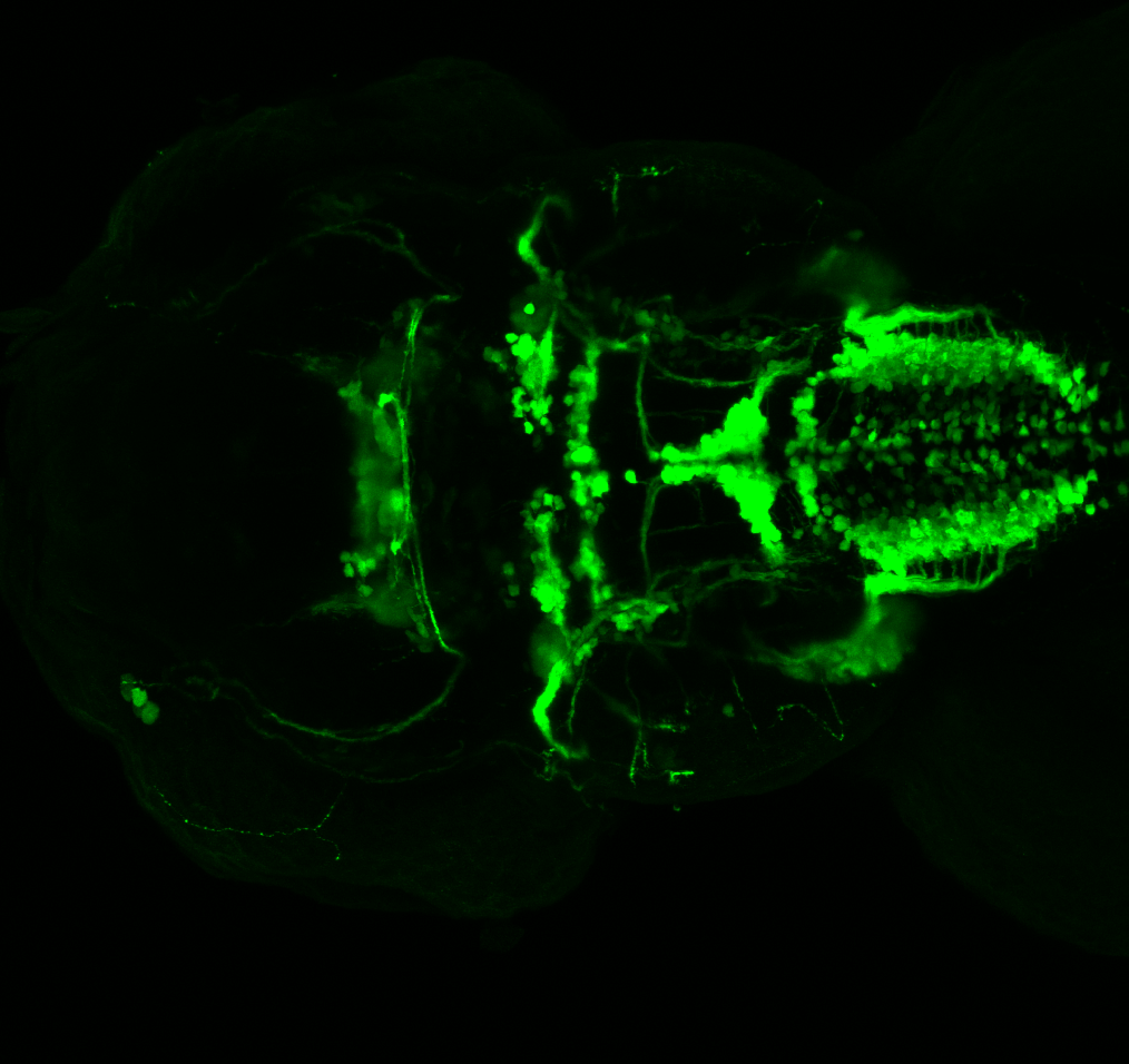
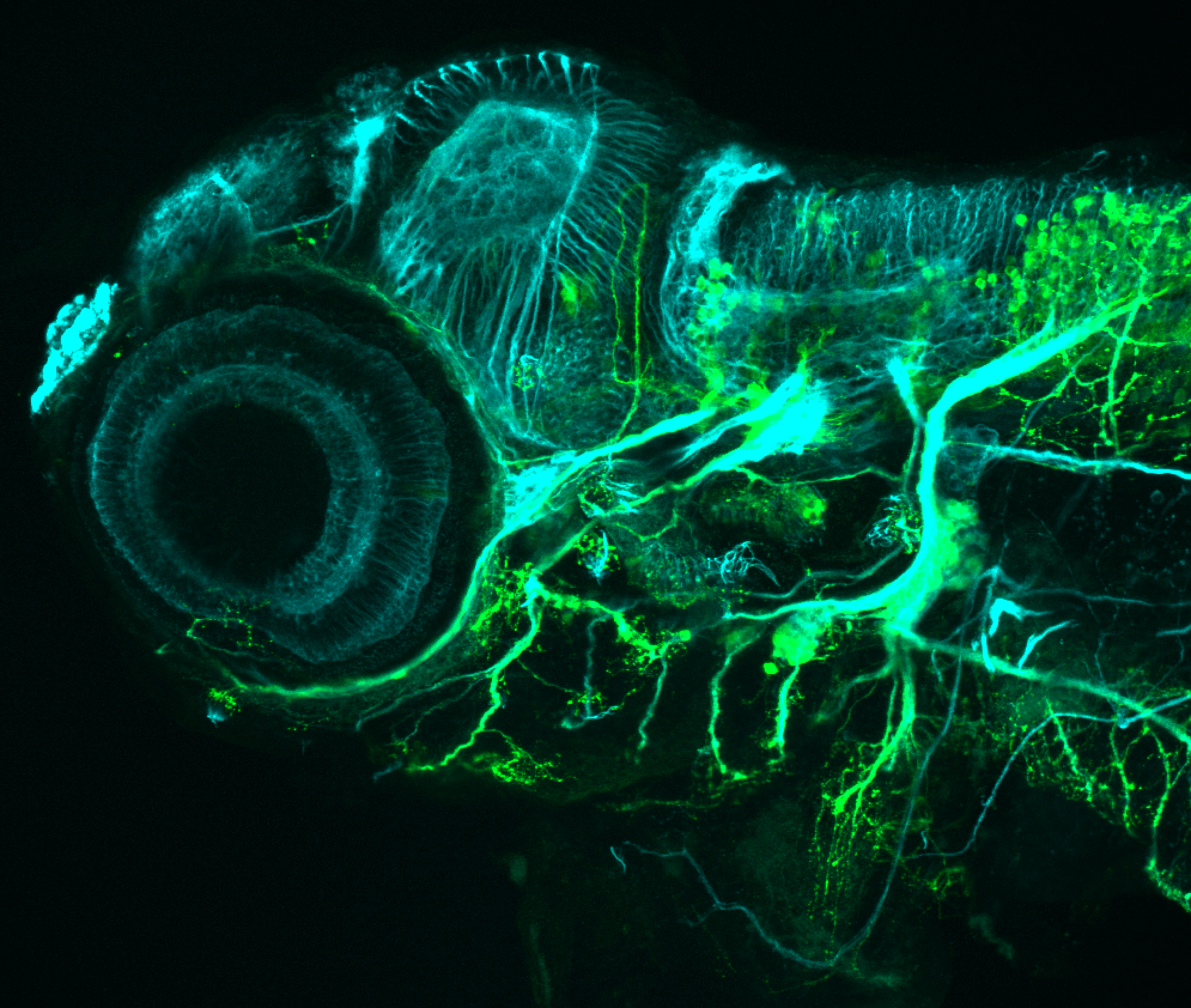
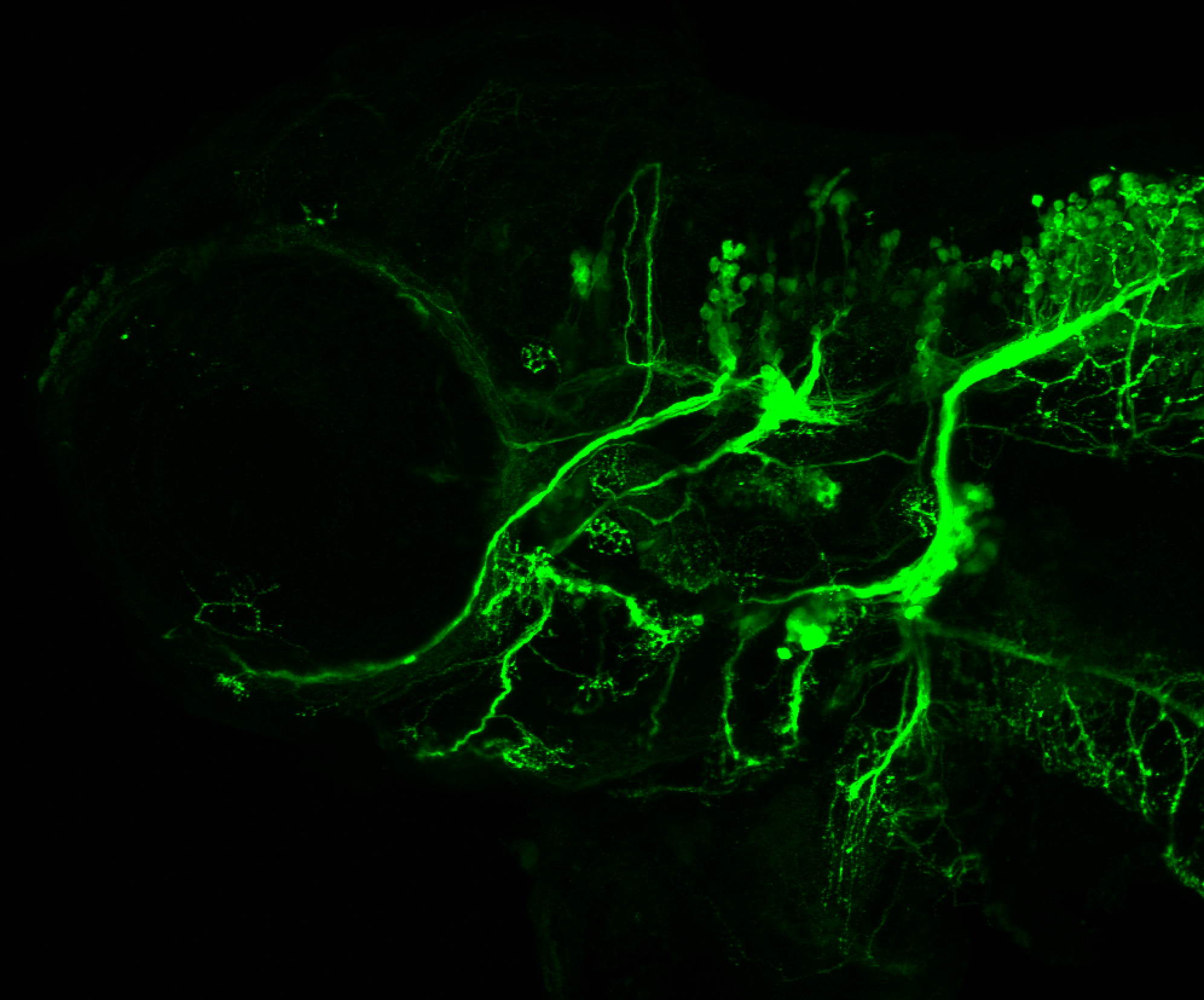
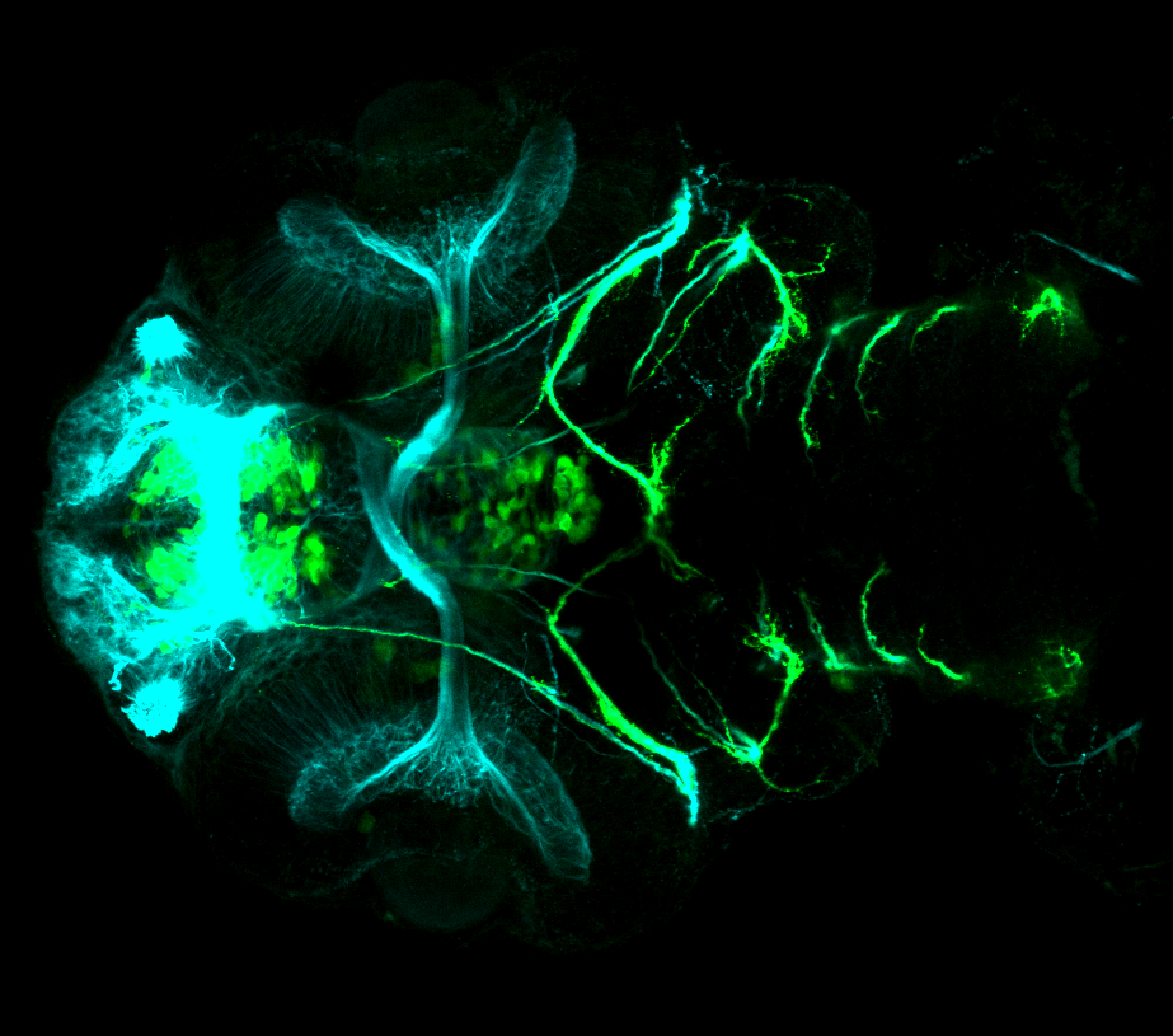
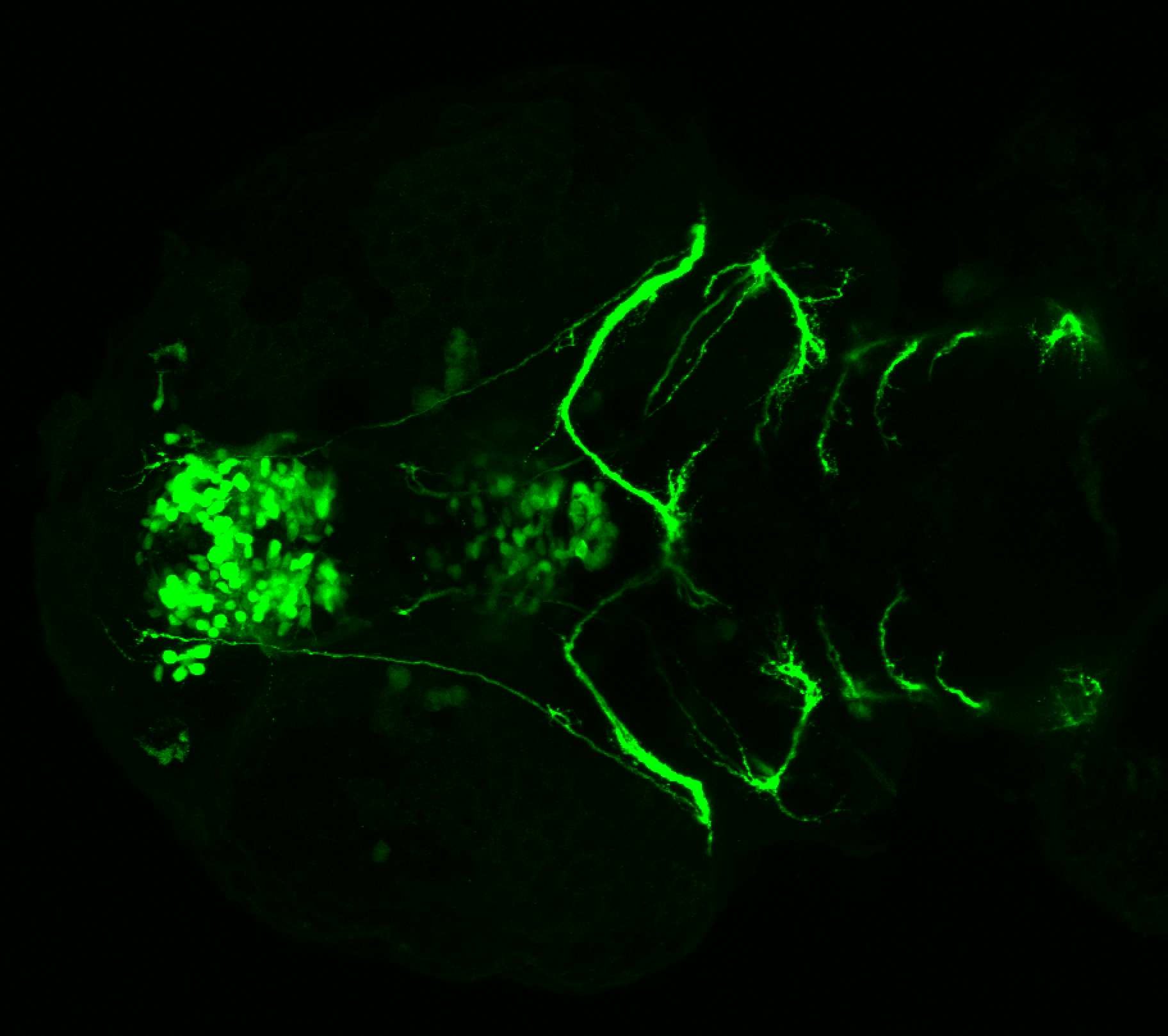
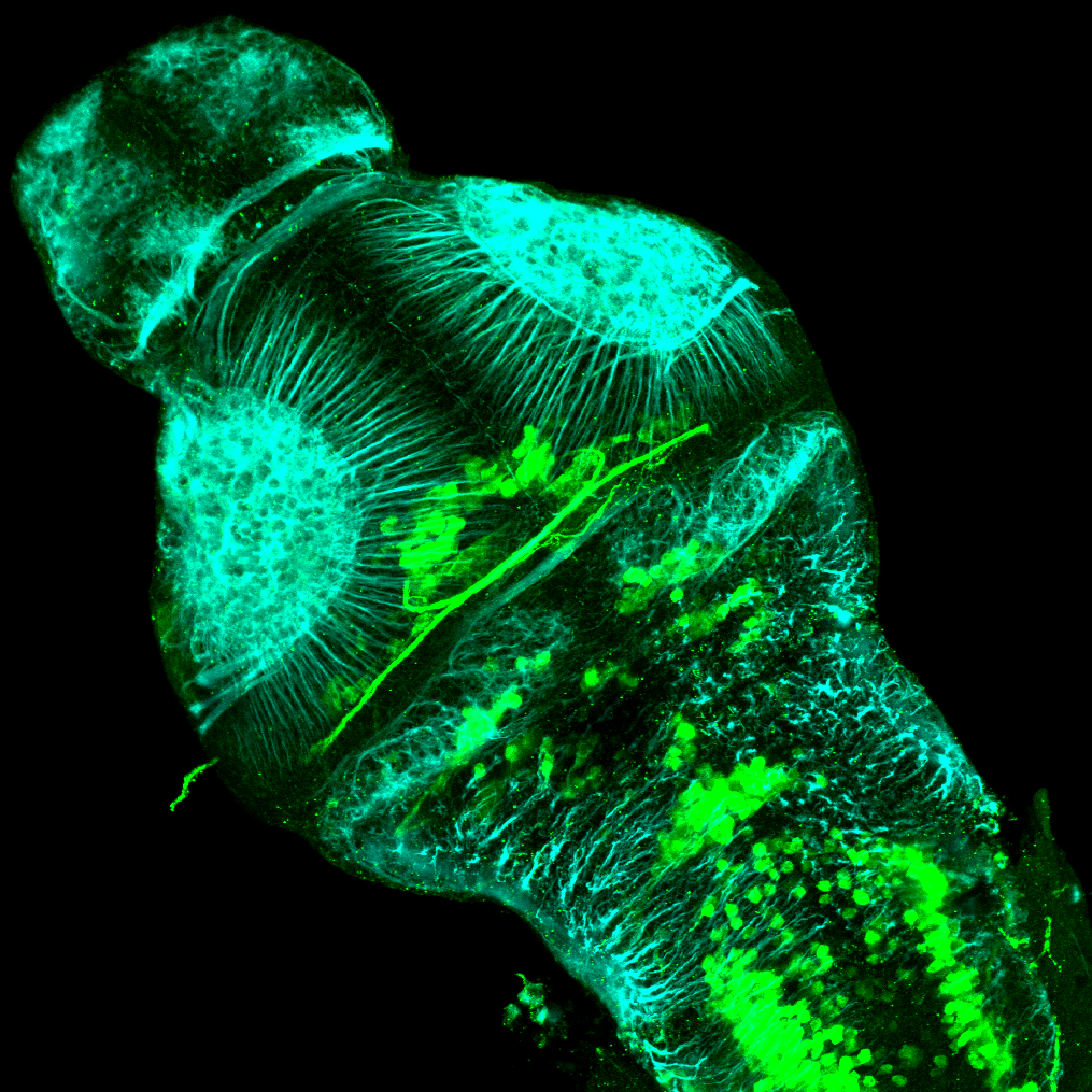
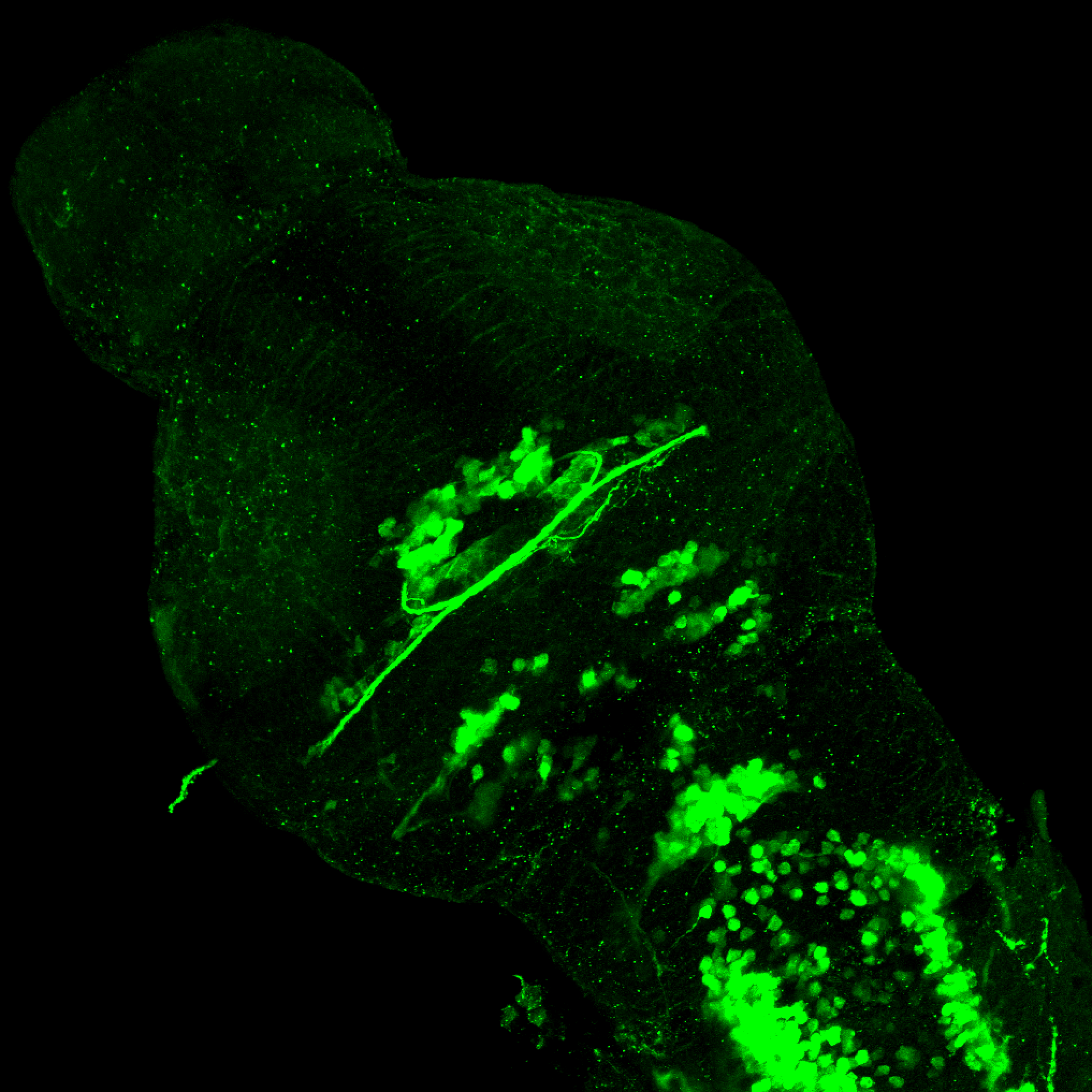
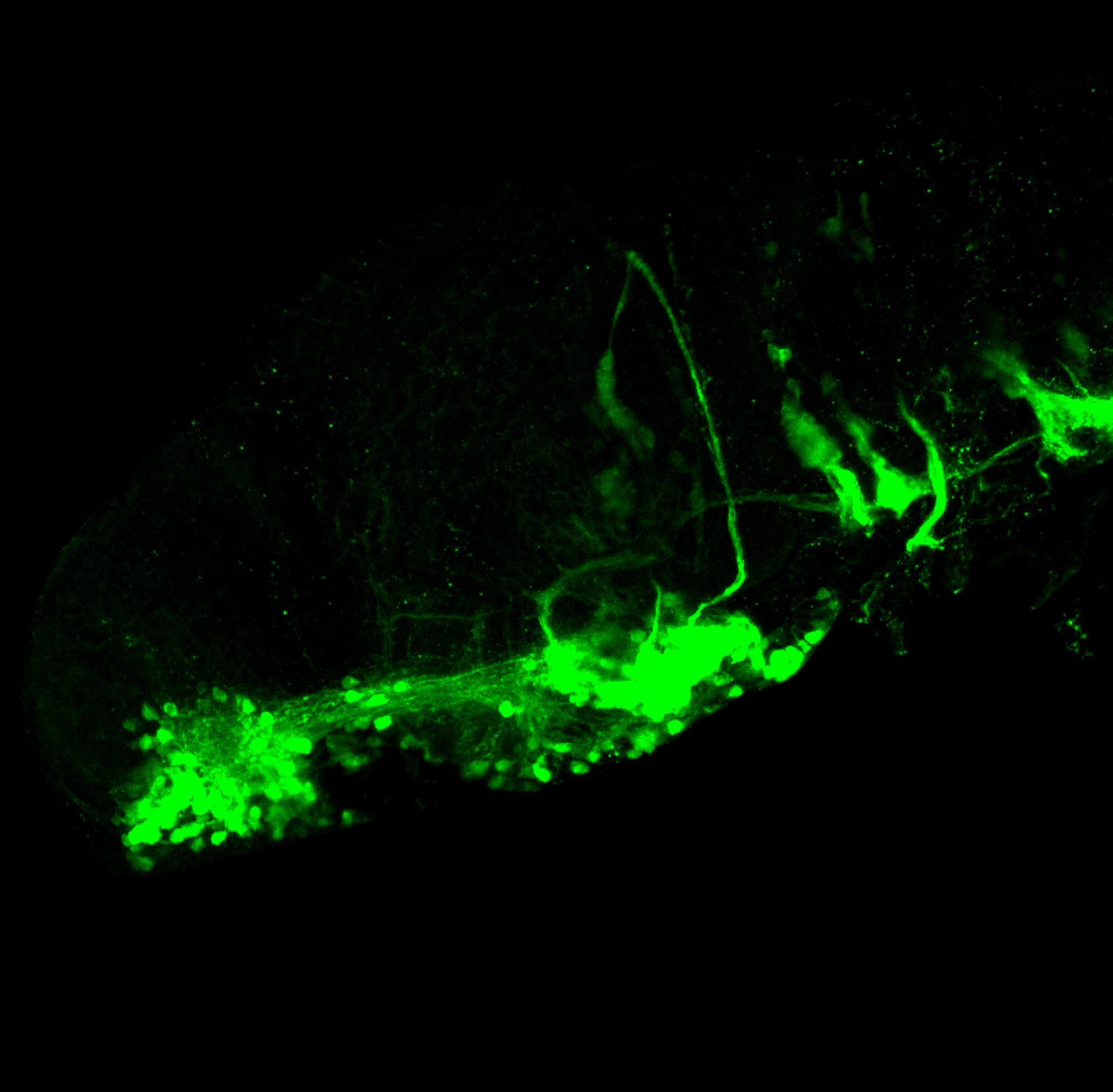
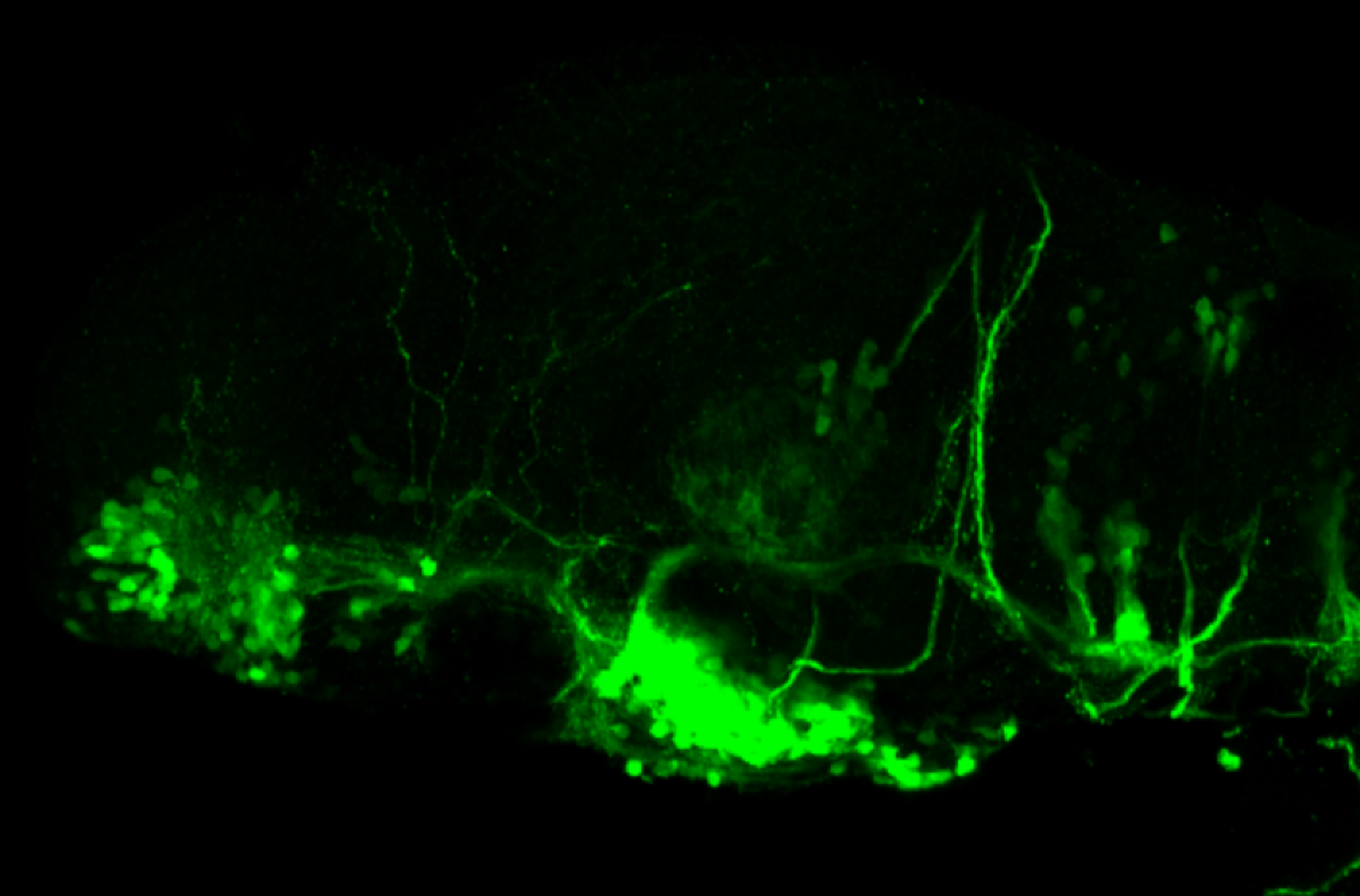
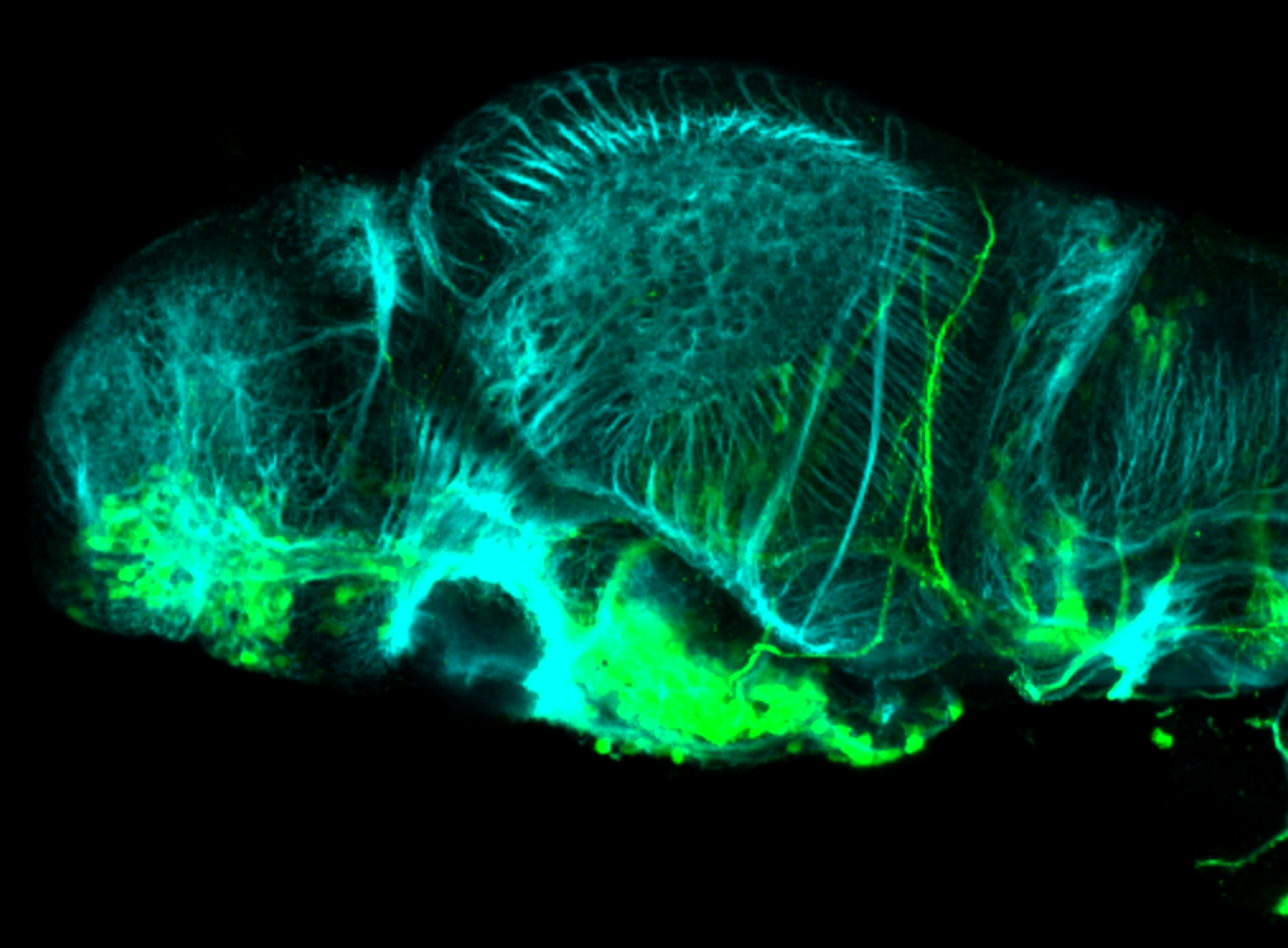
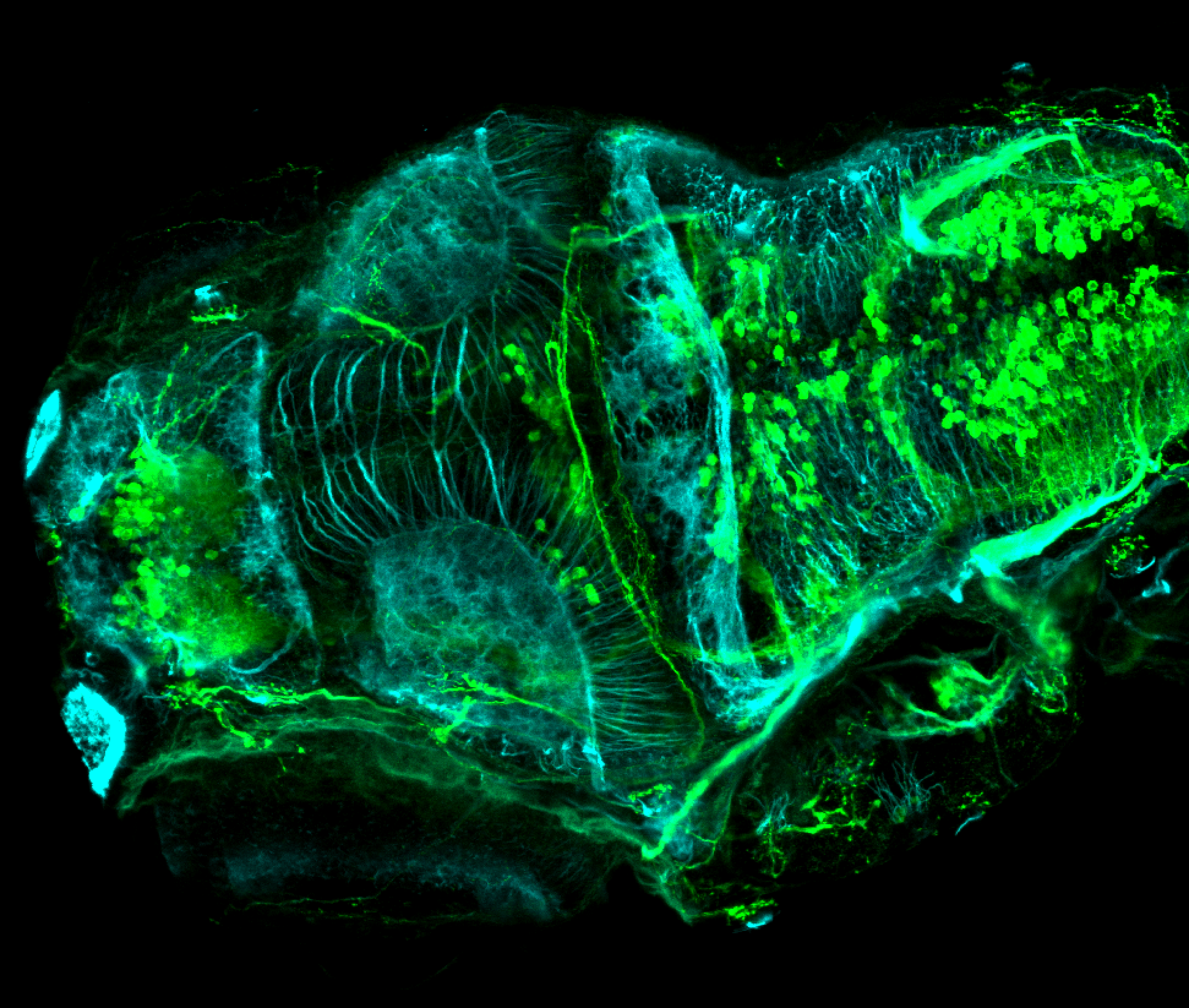
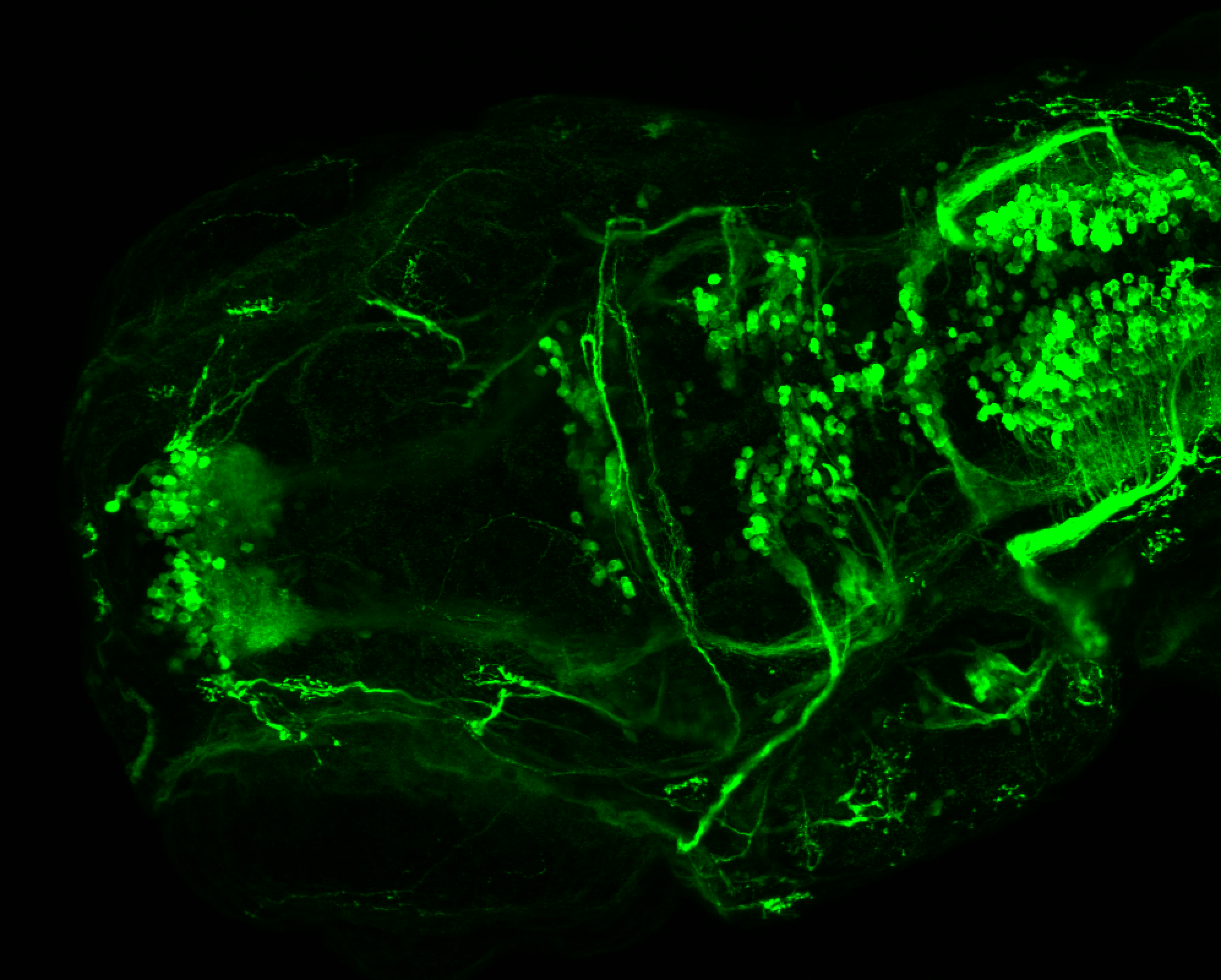
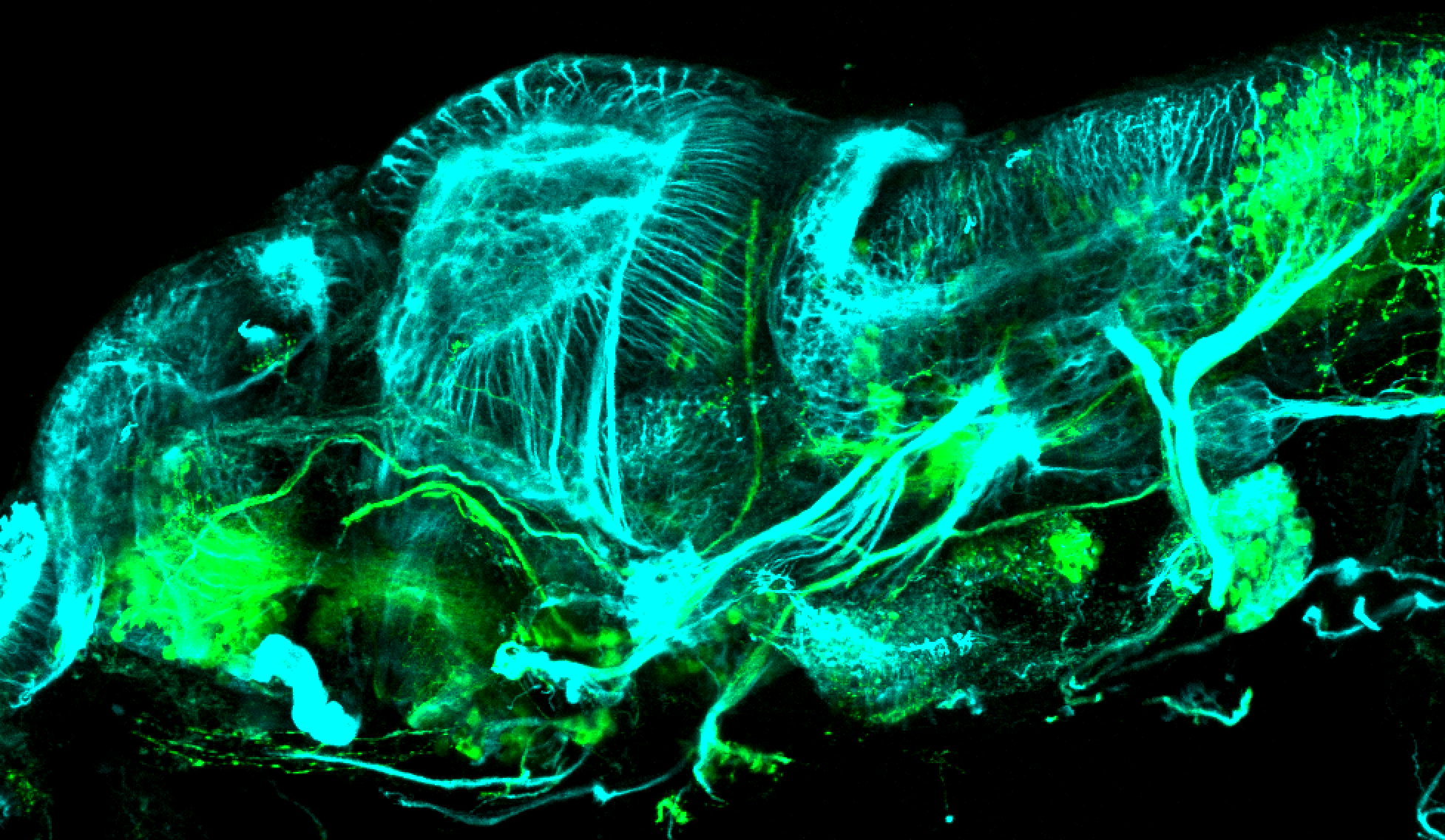
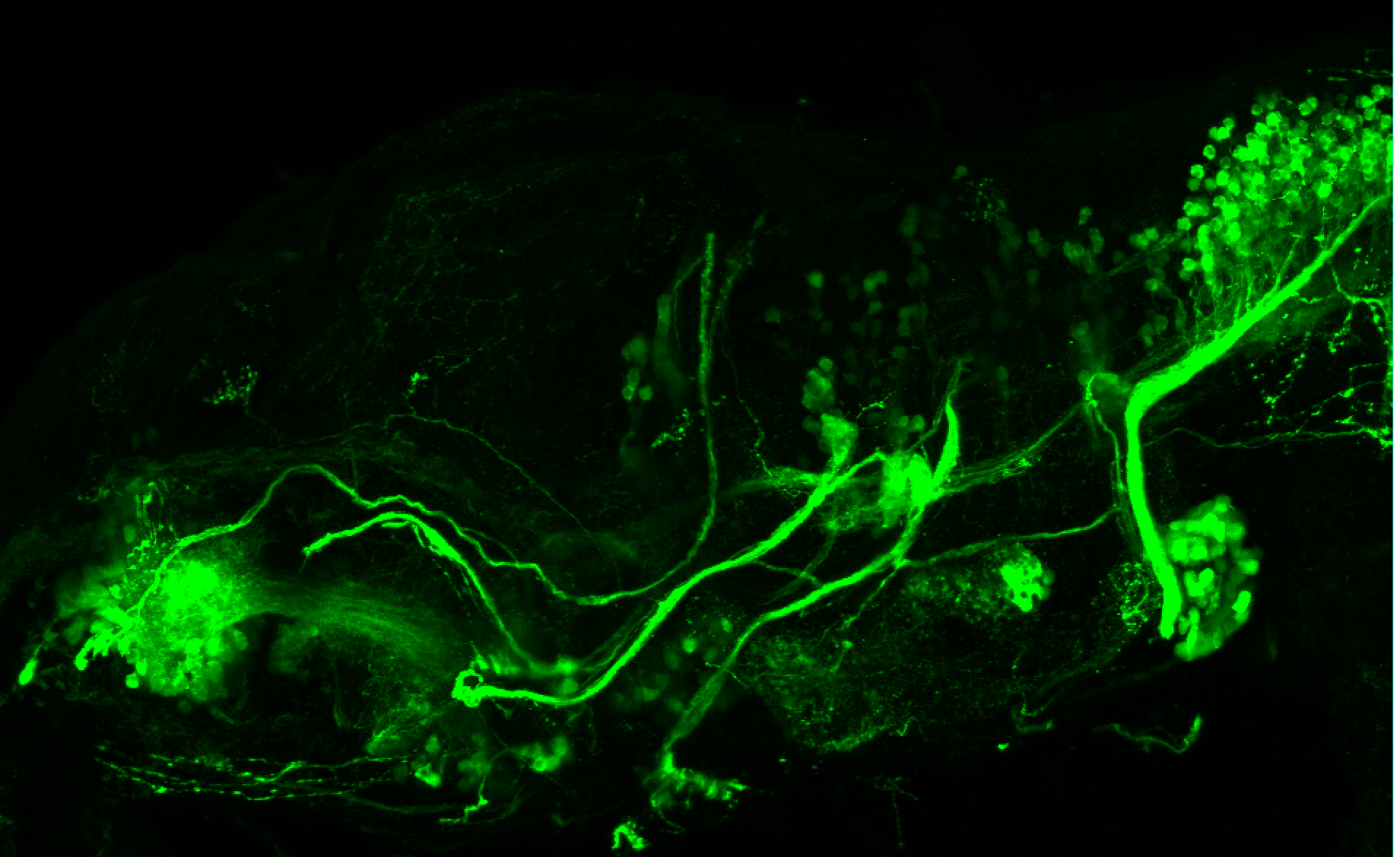
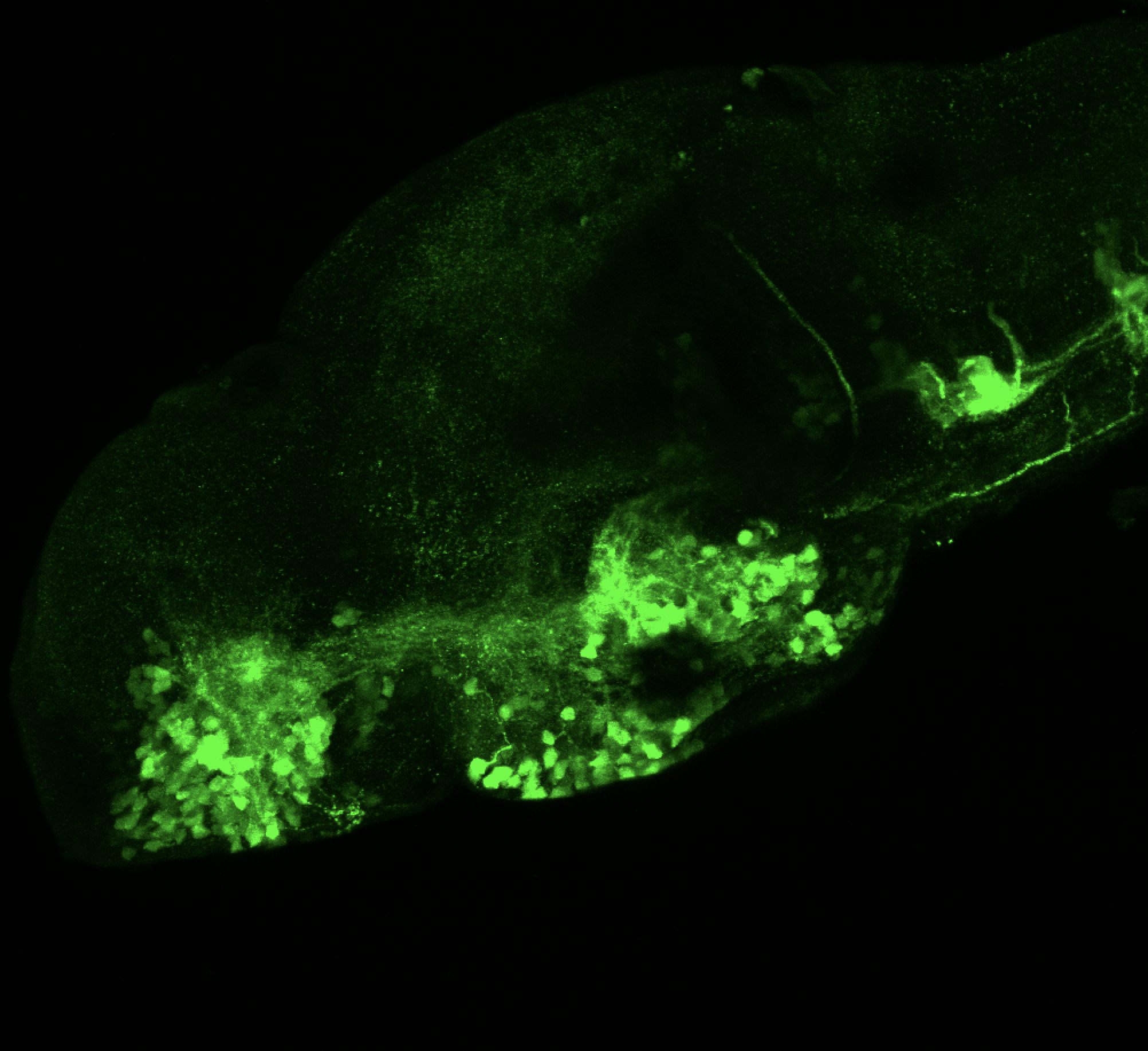
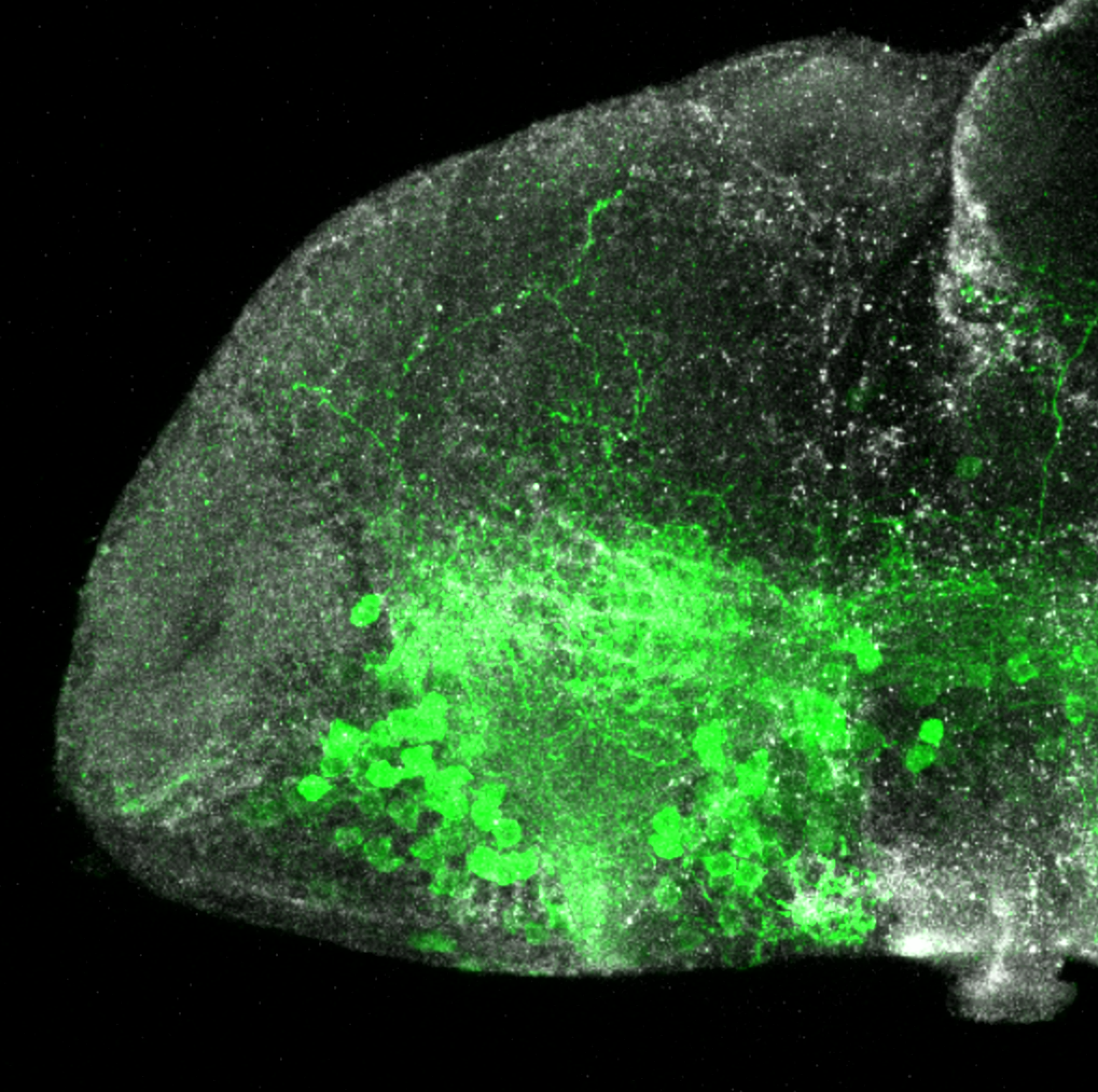
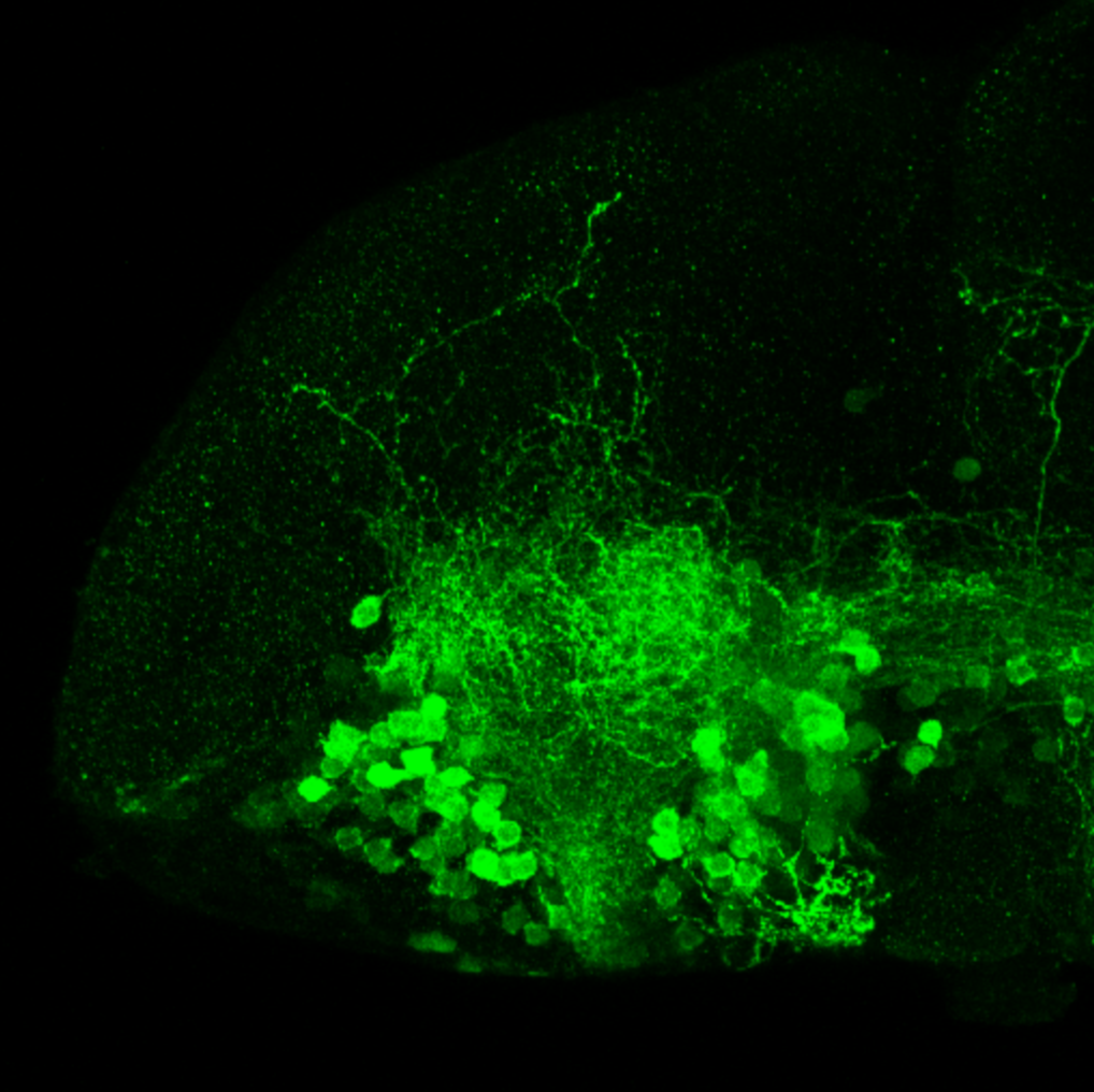
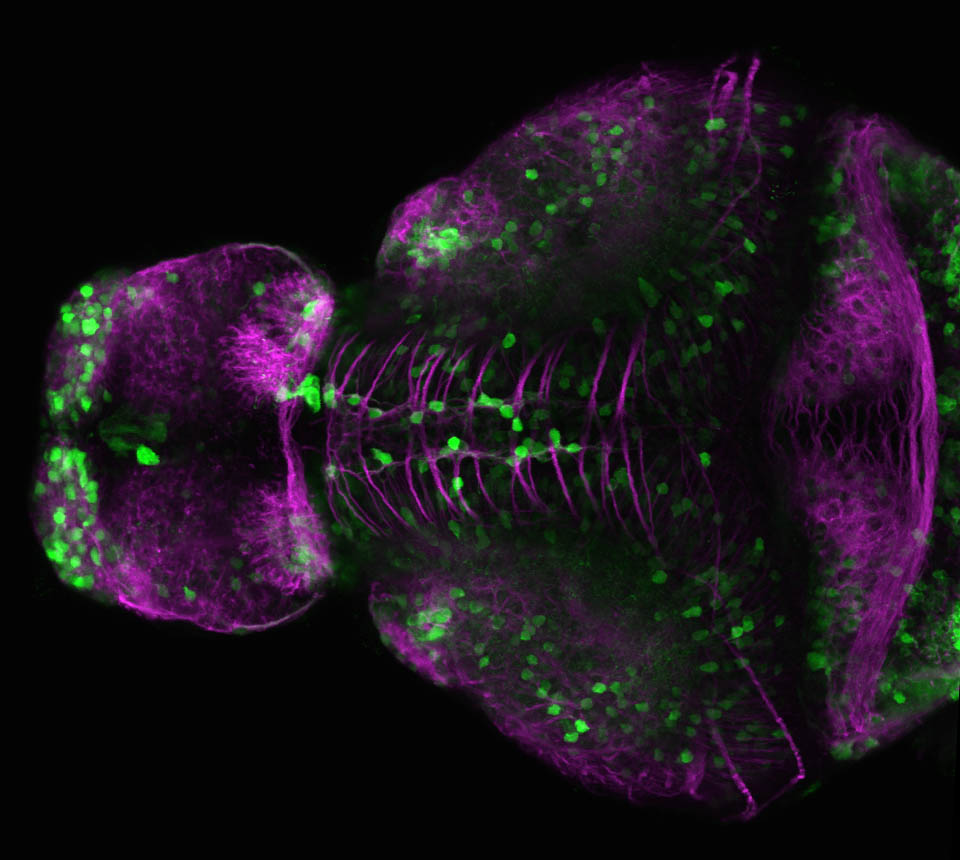
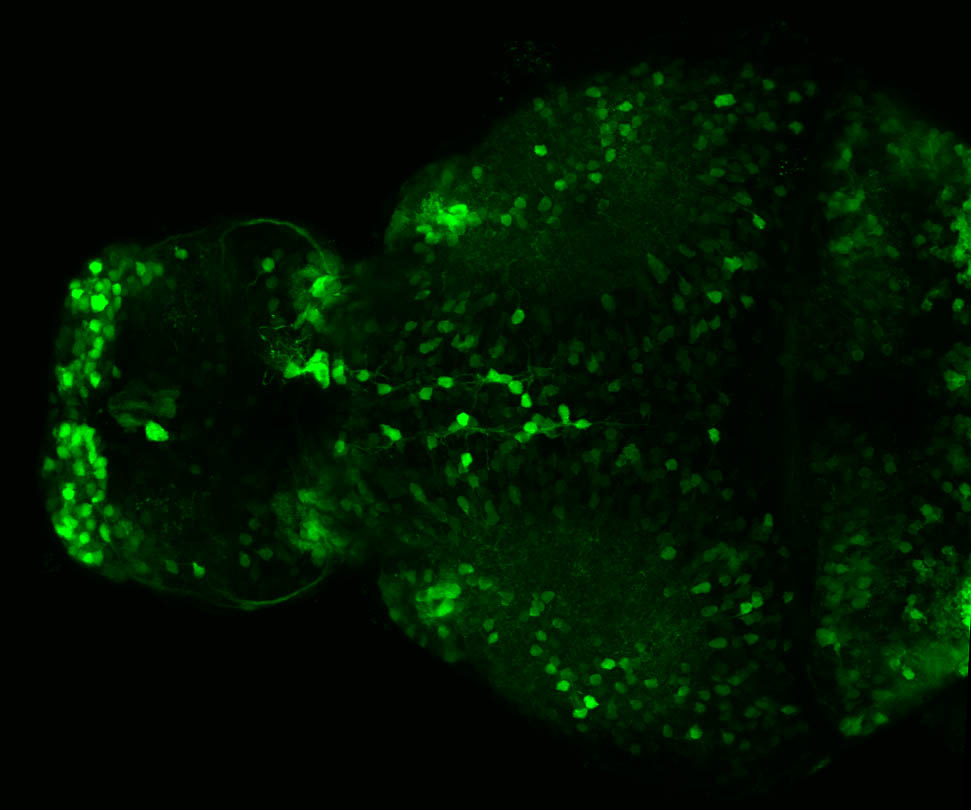
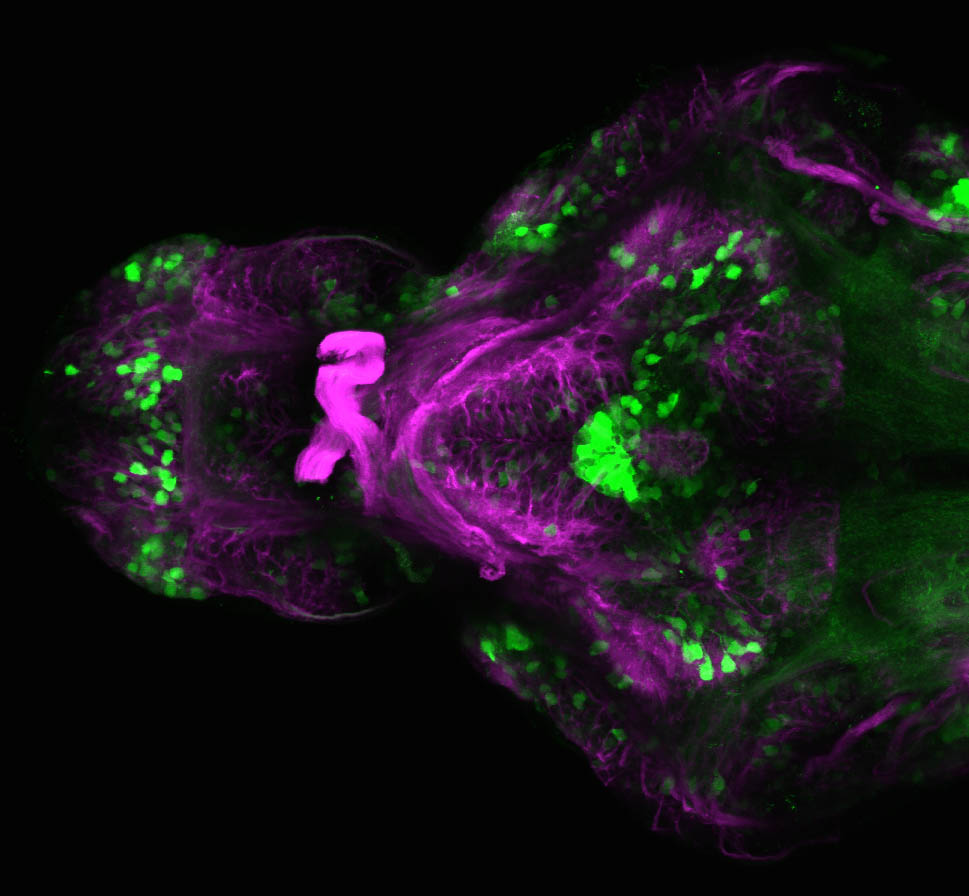
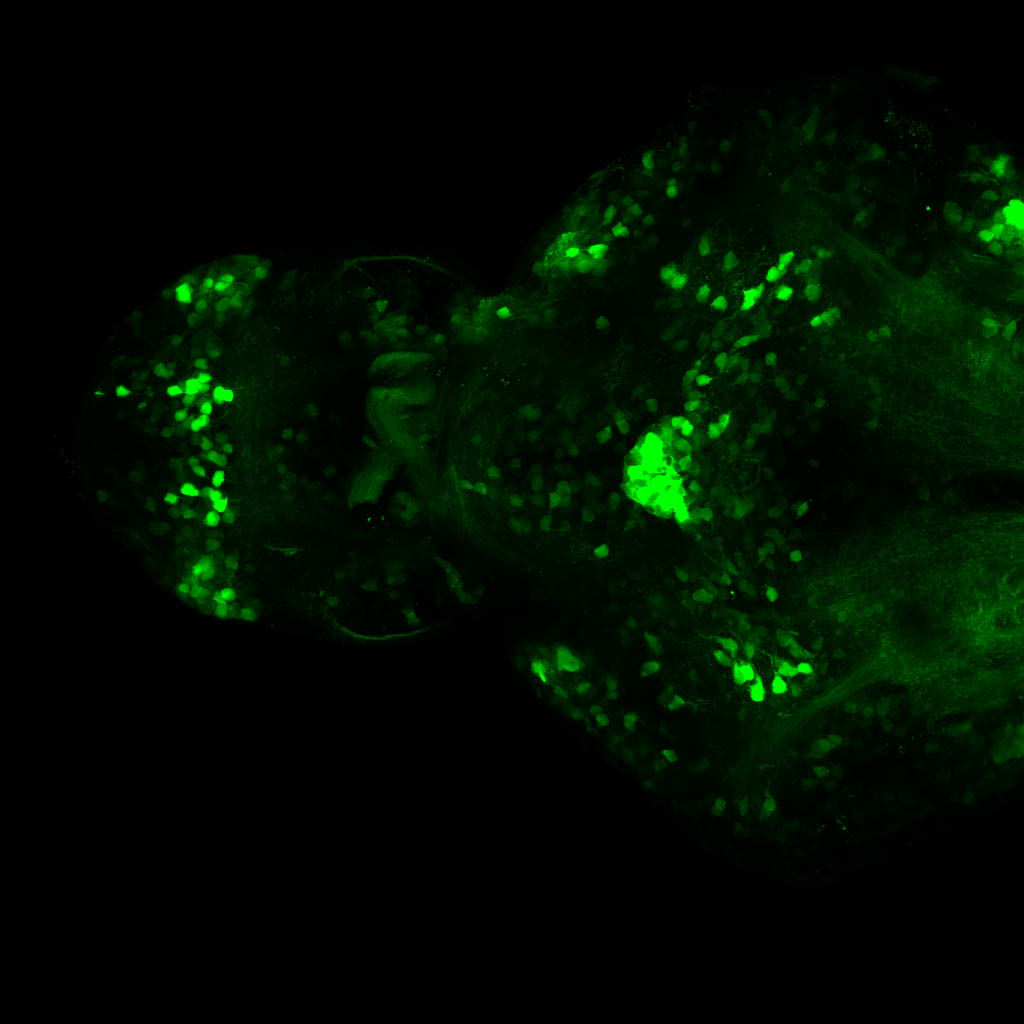
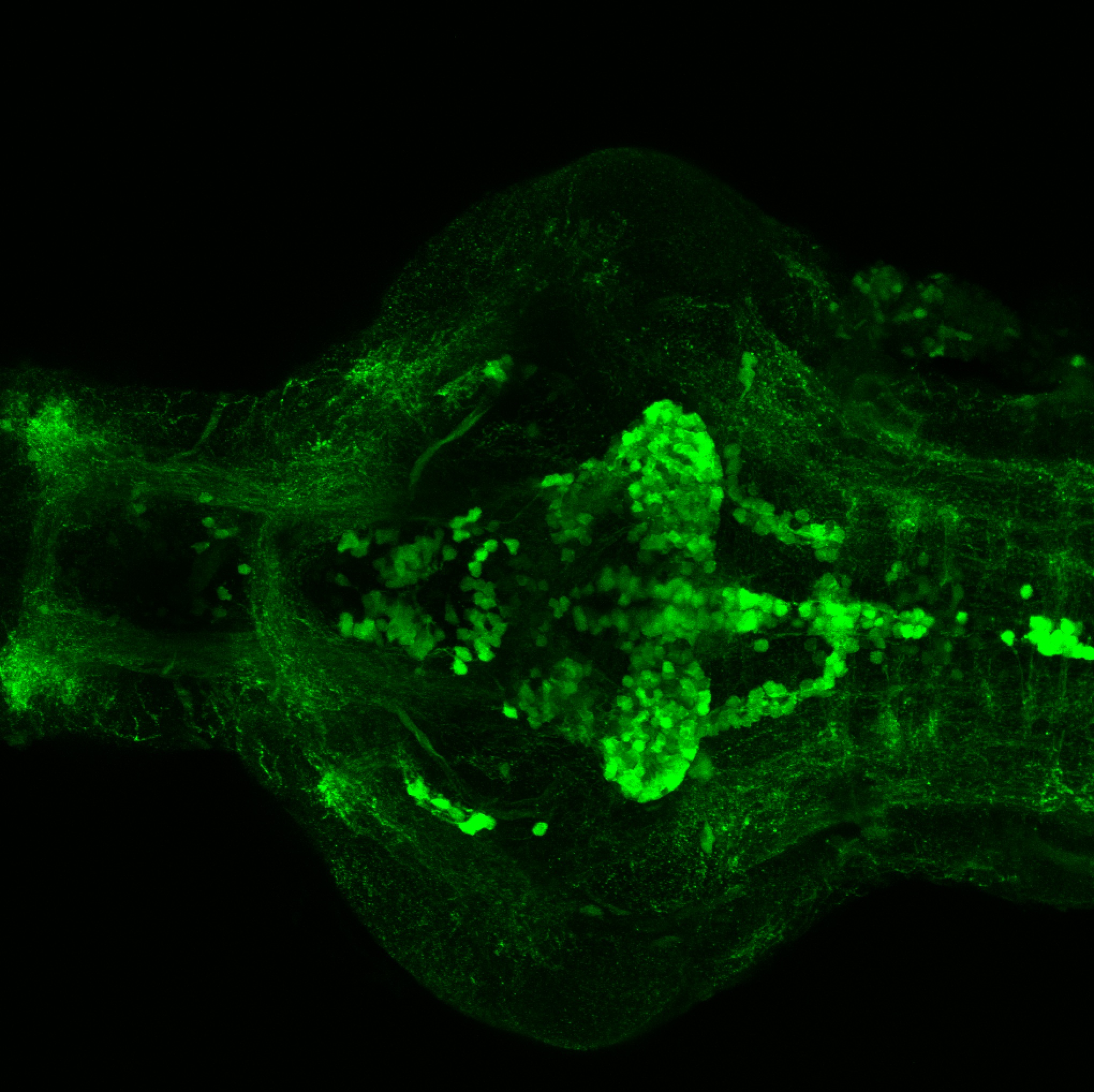
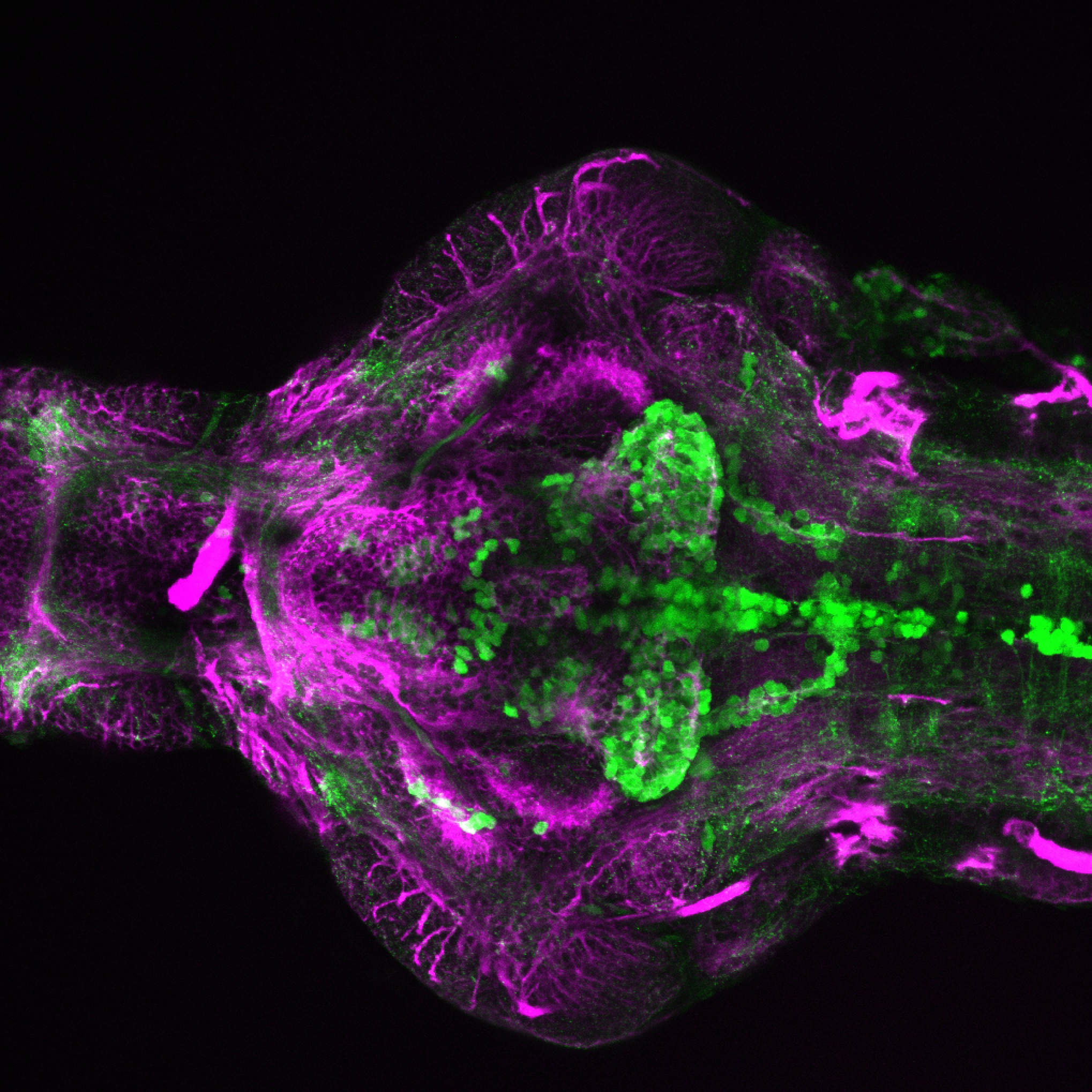
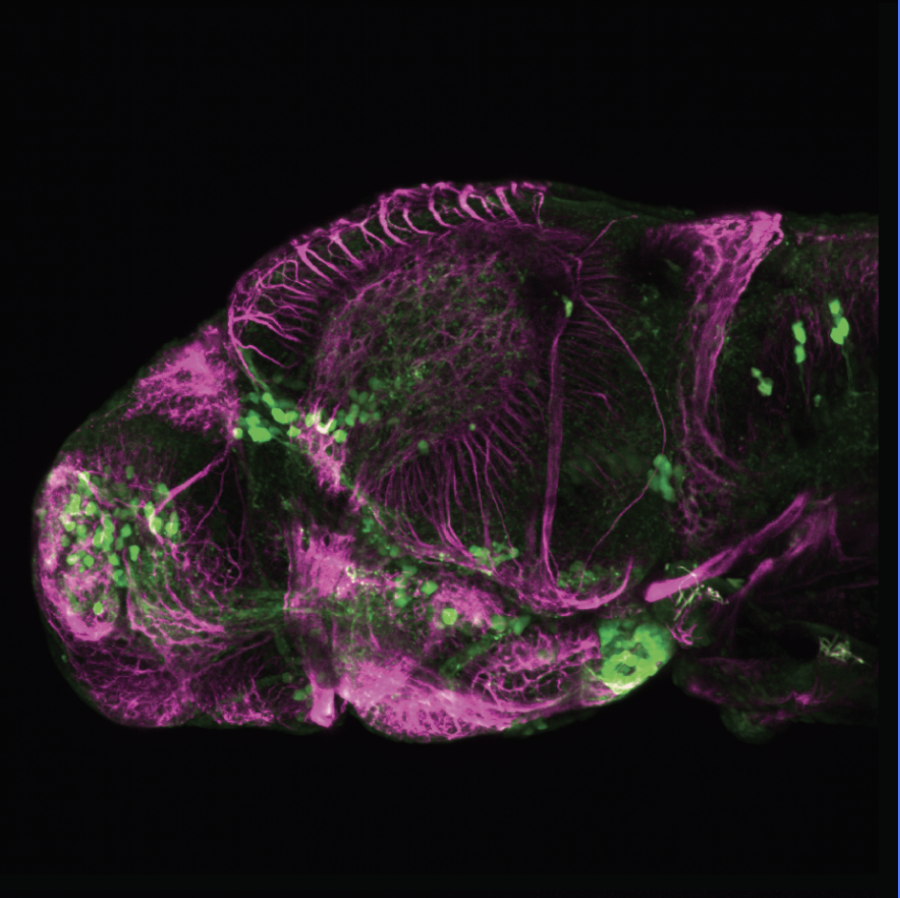
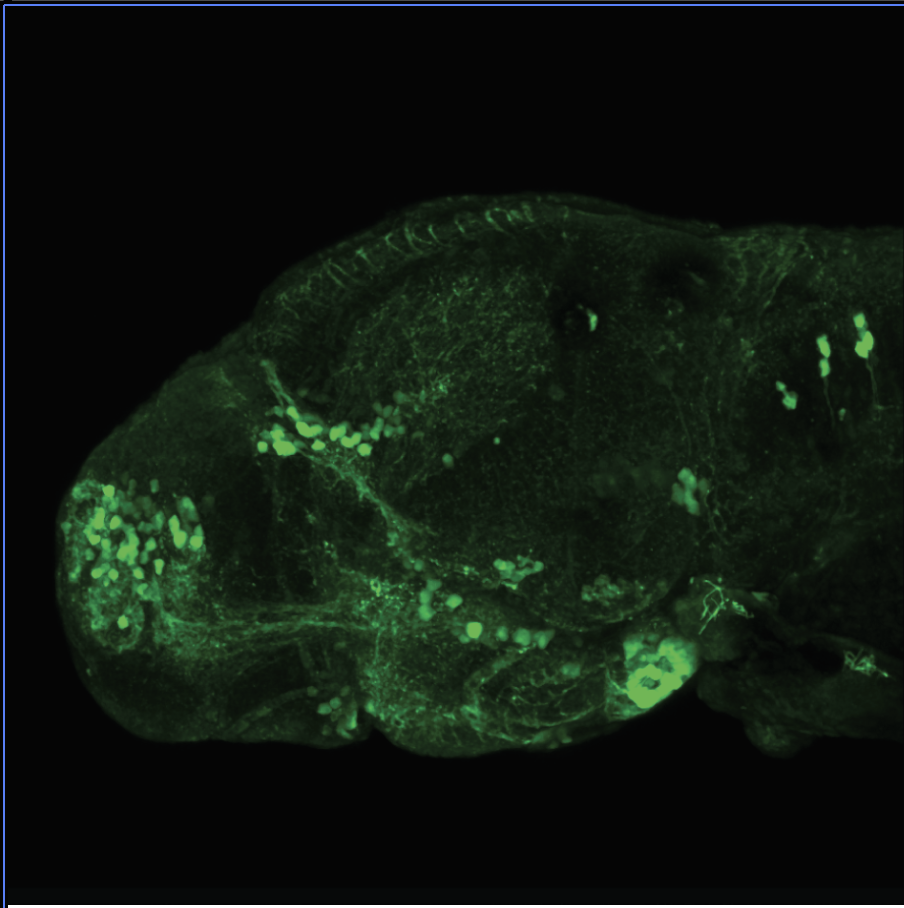
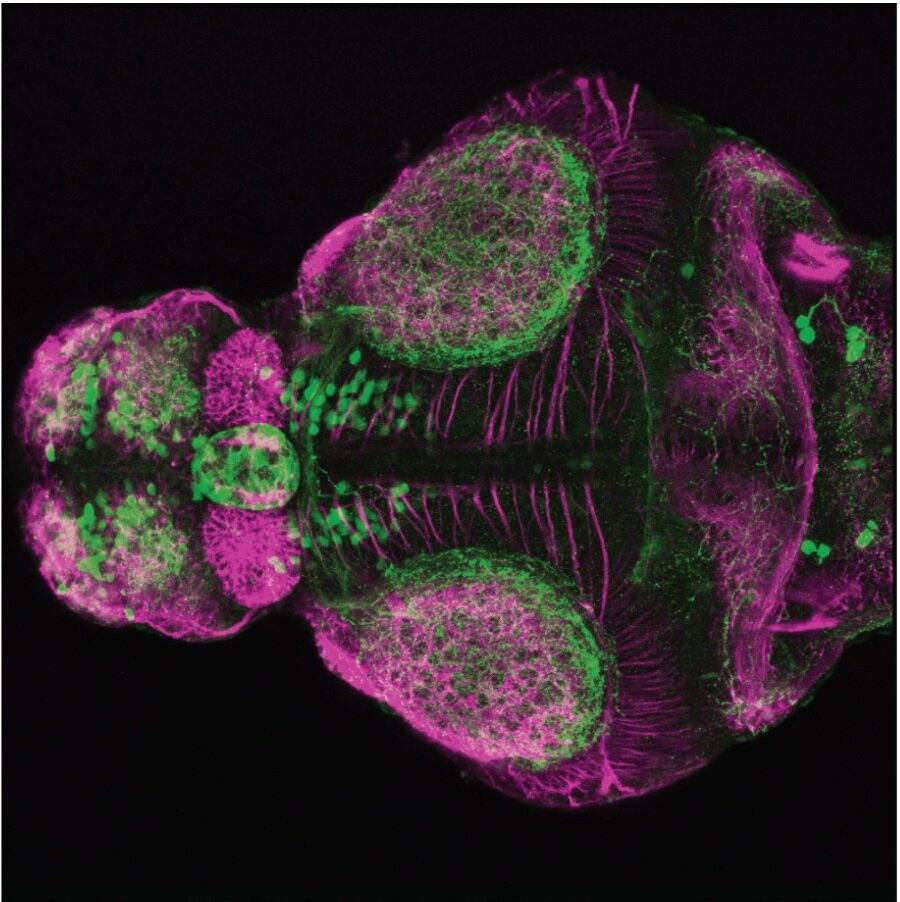
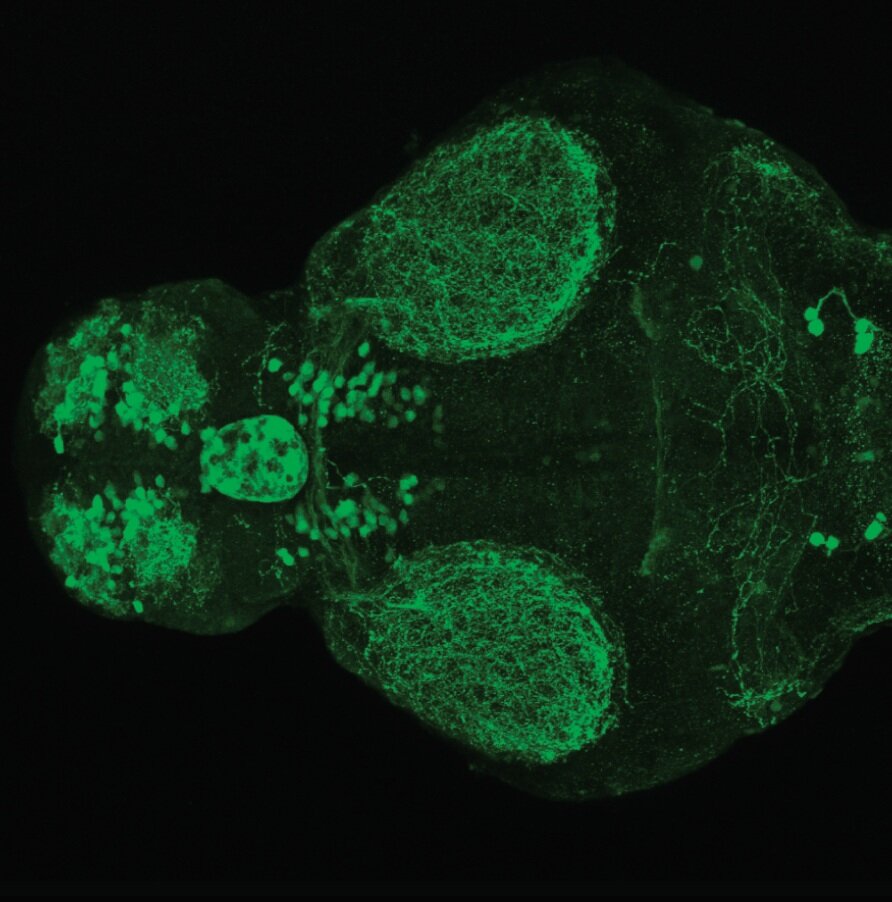
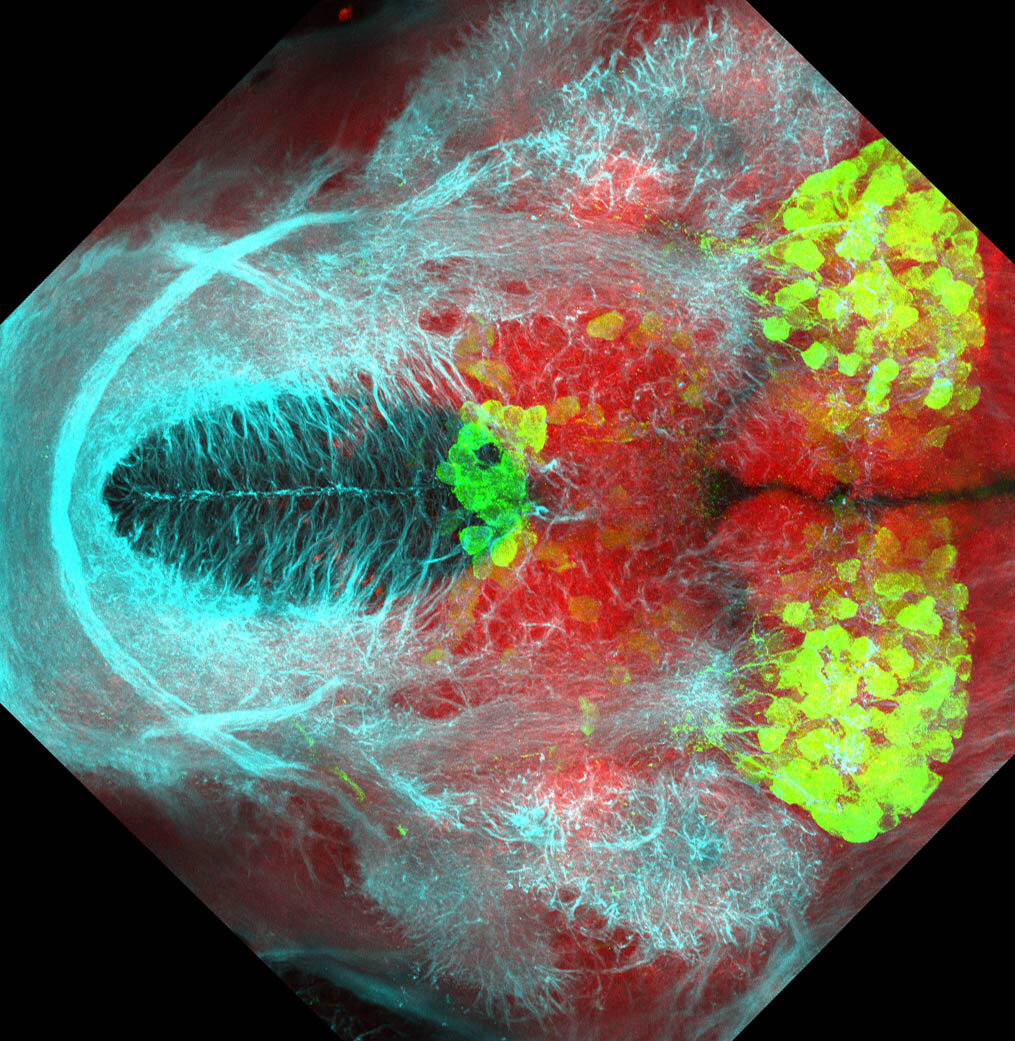
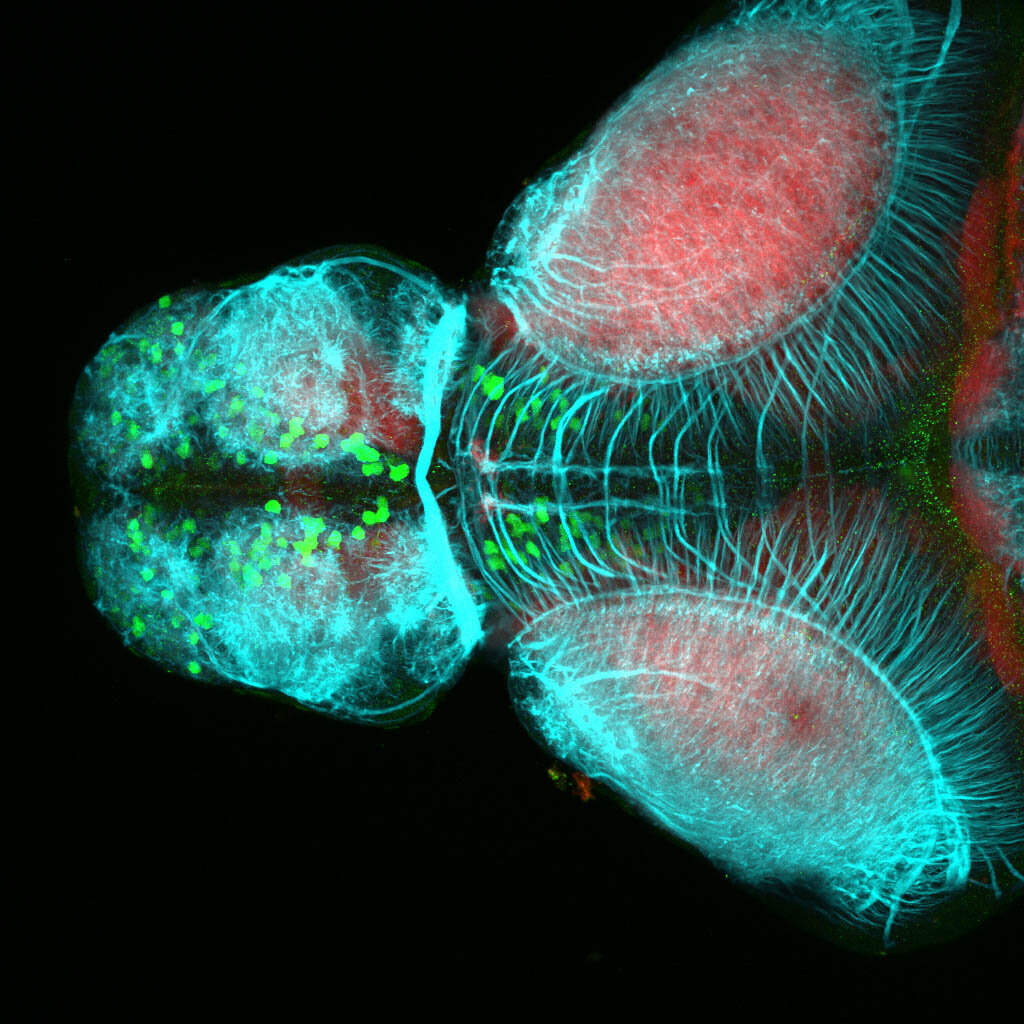
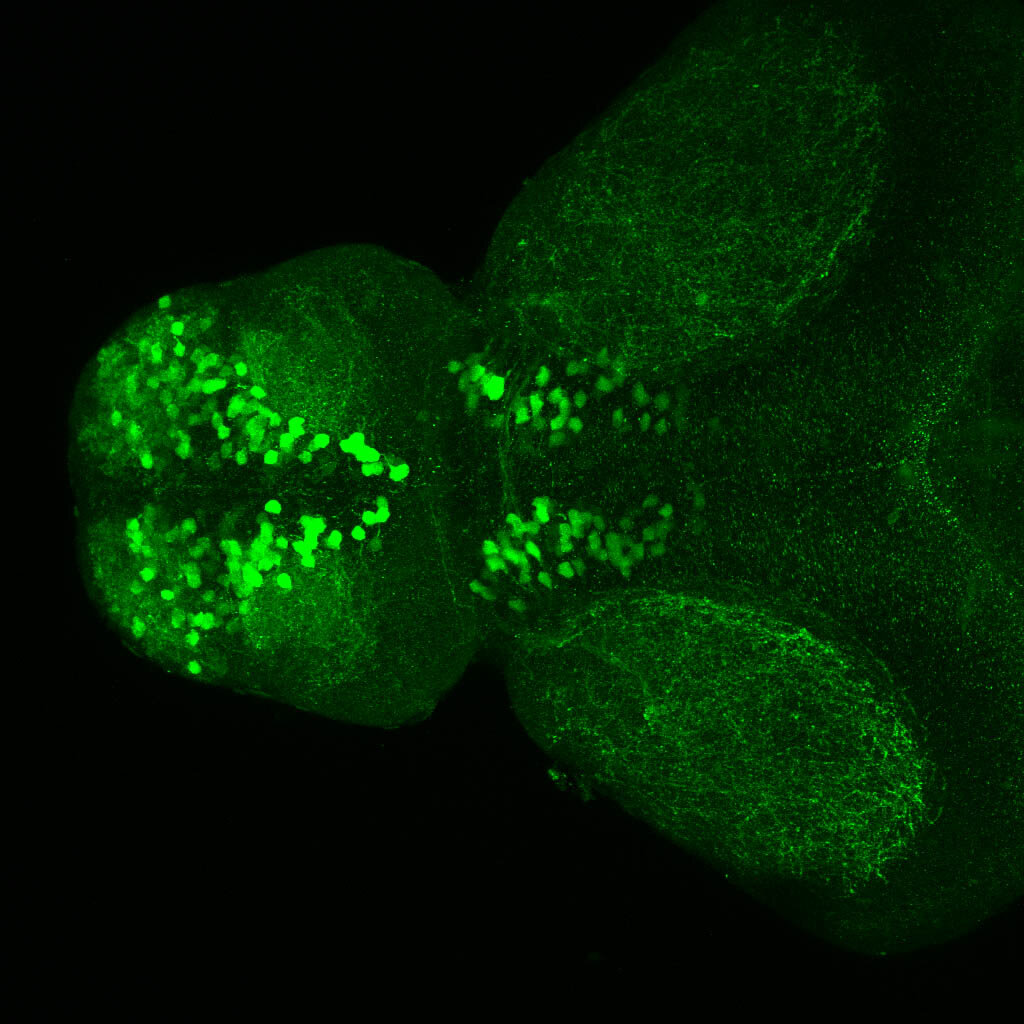
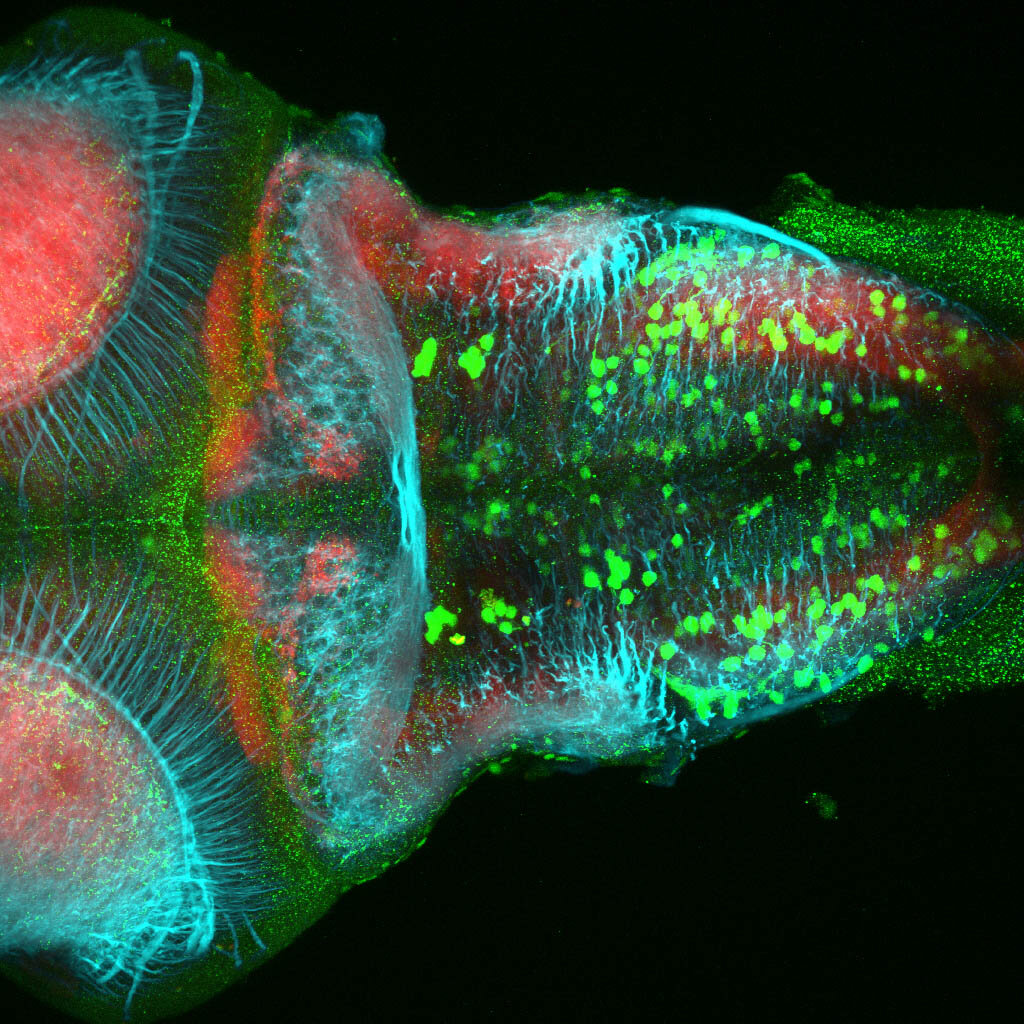
The Gilmore lab wanted to label the lateral line and neuromasts of the lateral line system, one allele of this transgenic also had EGFP expression in the nasal retina and telencephalon and has been used by other labs to study eye and telencephalic morphogenesis.
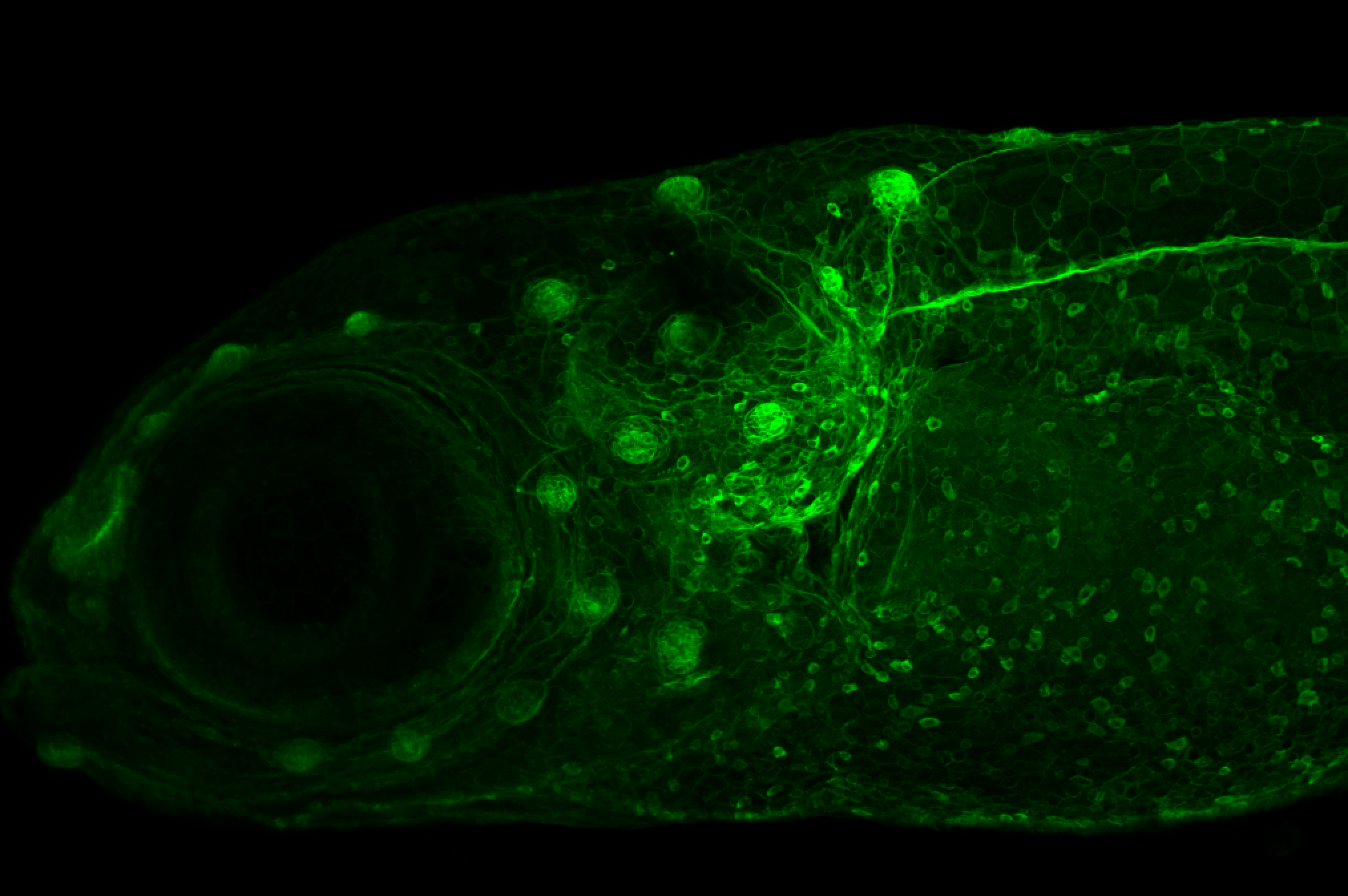
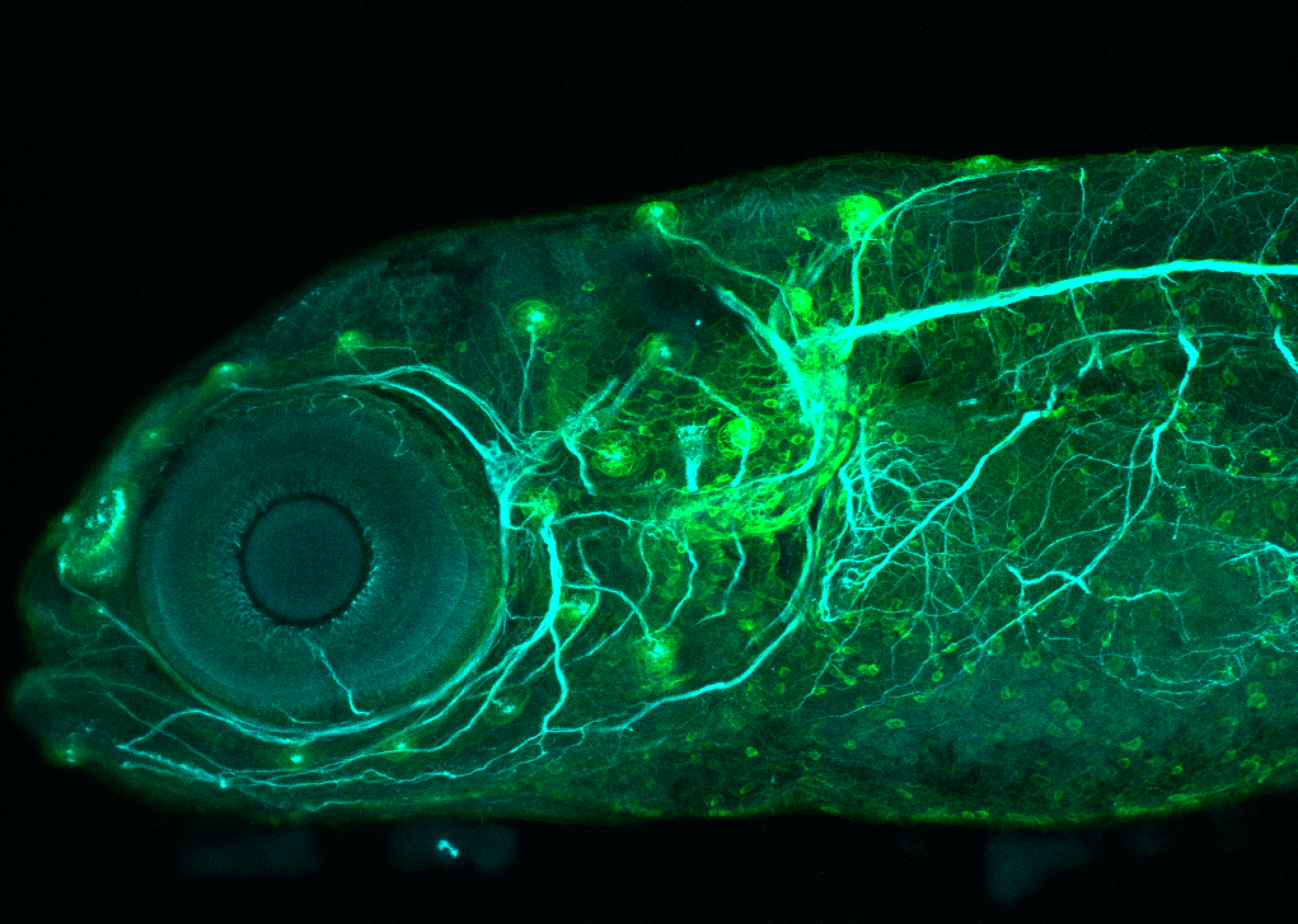
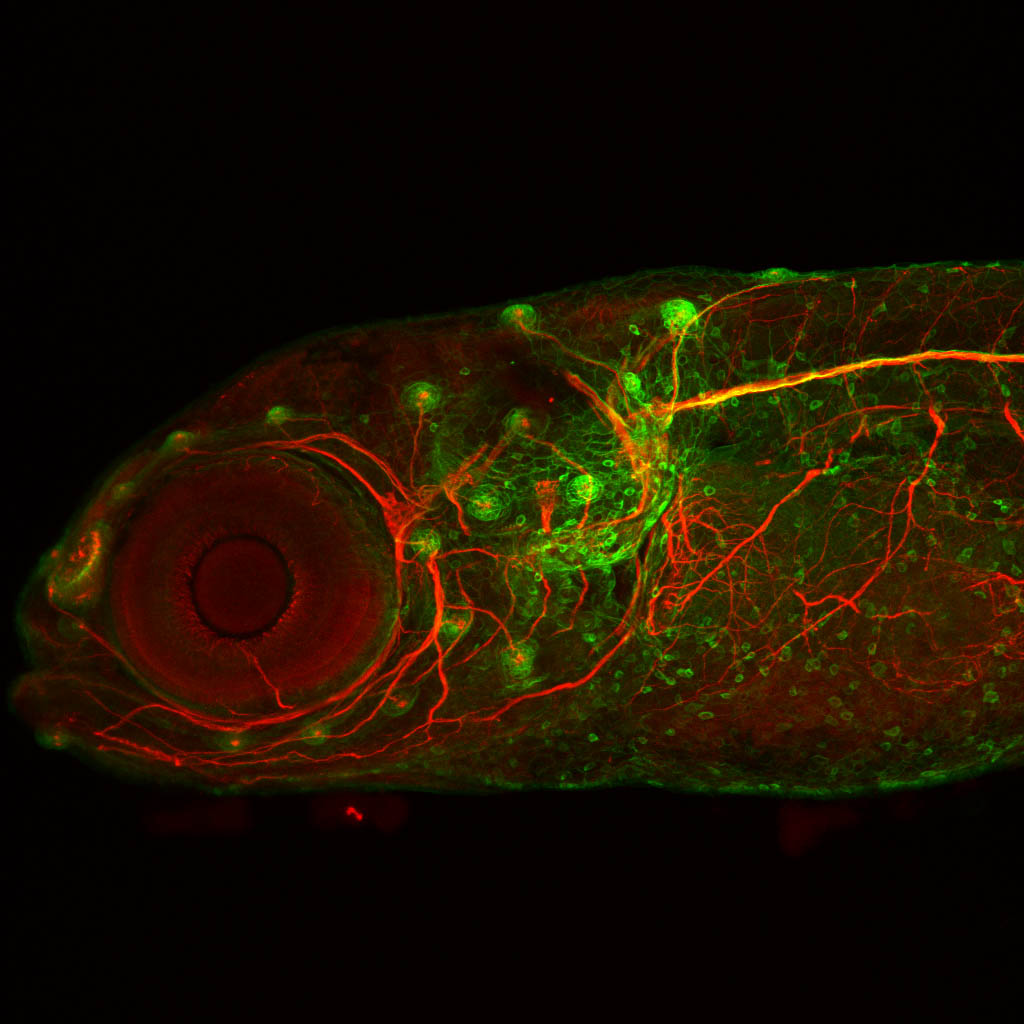
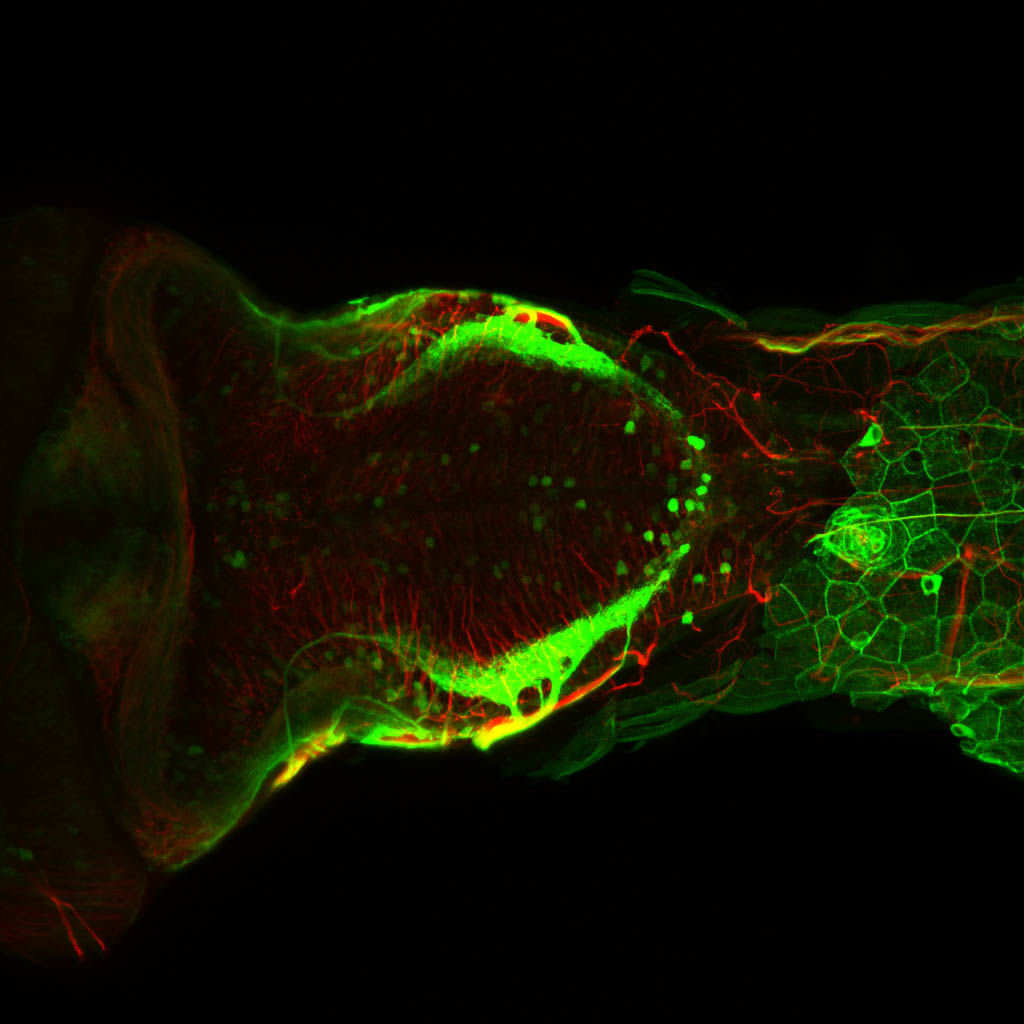
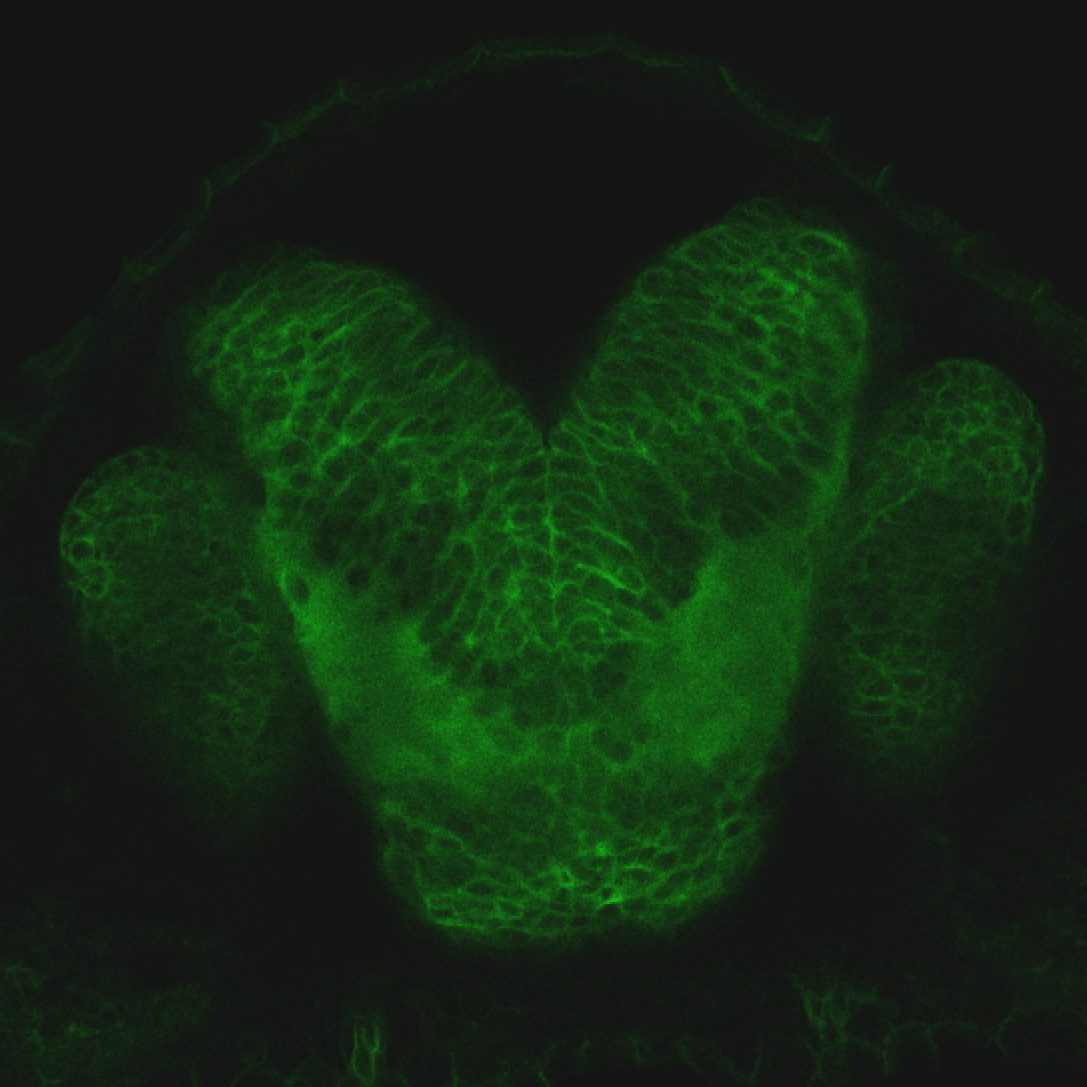
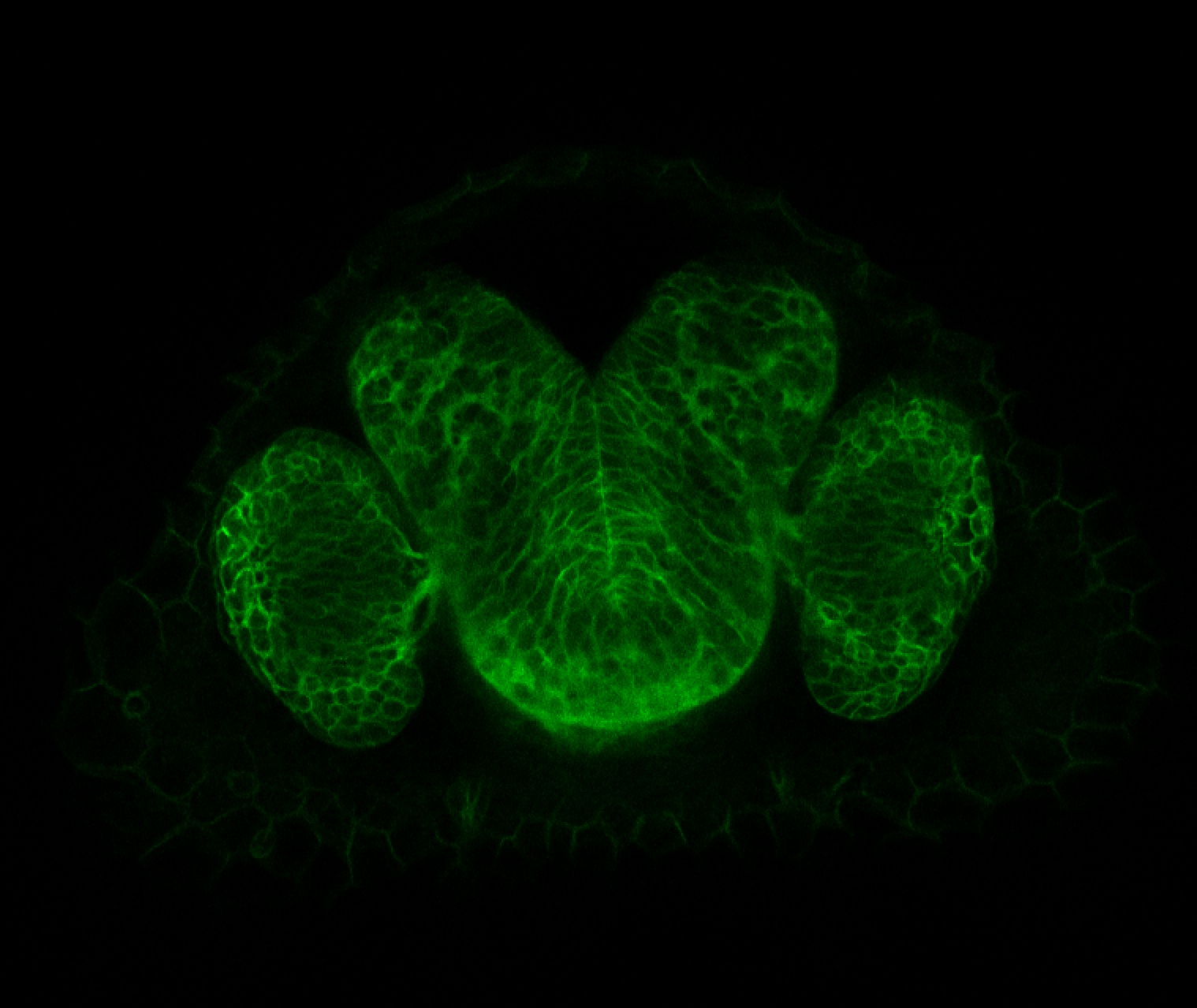
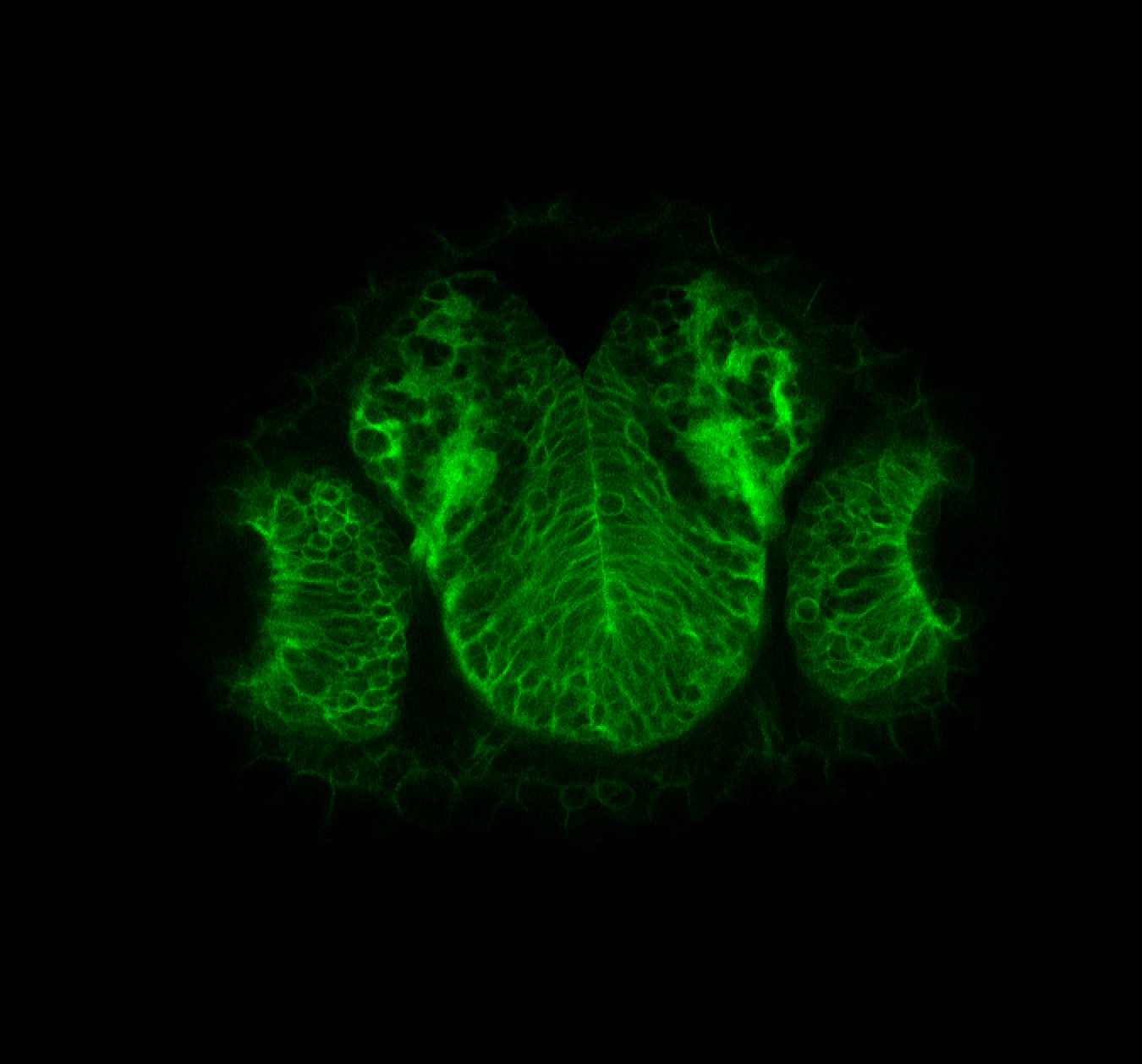
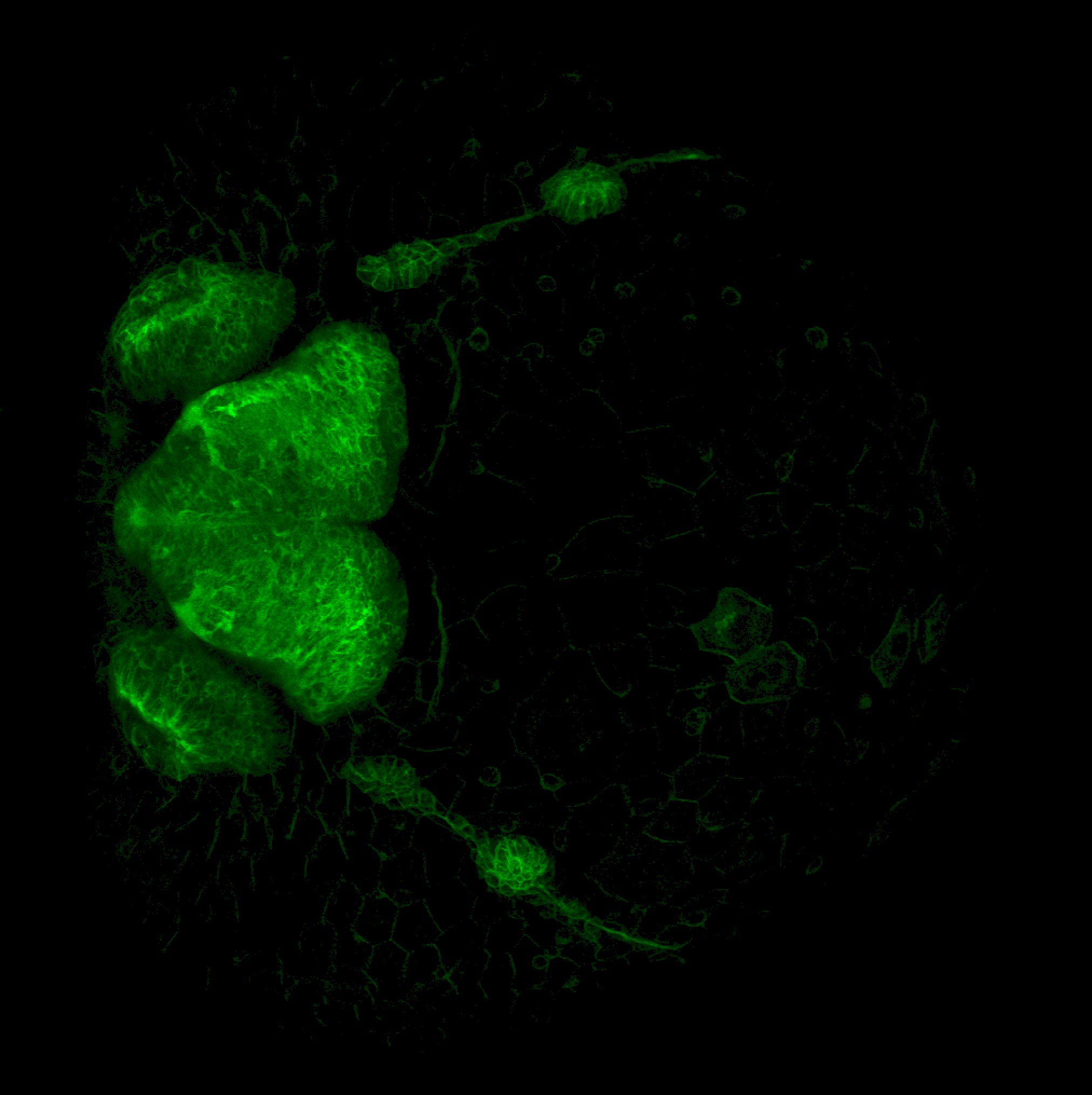
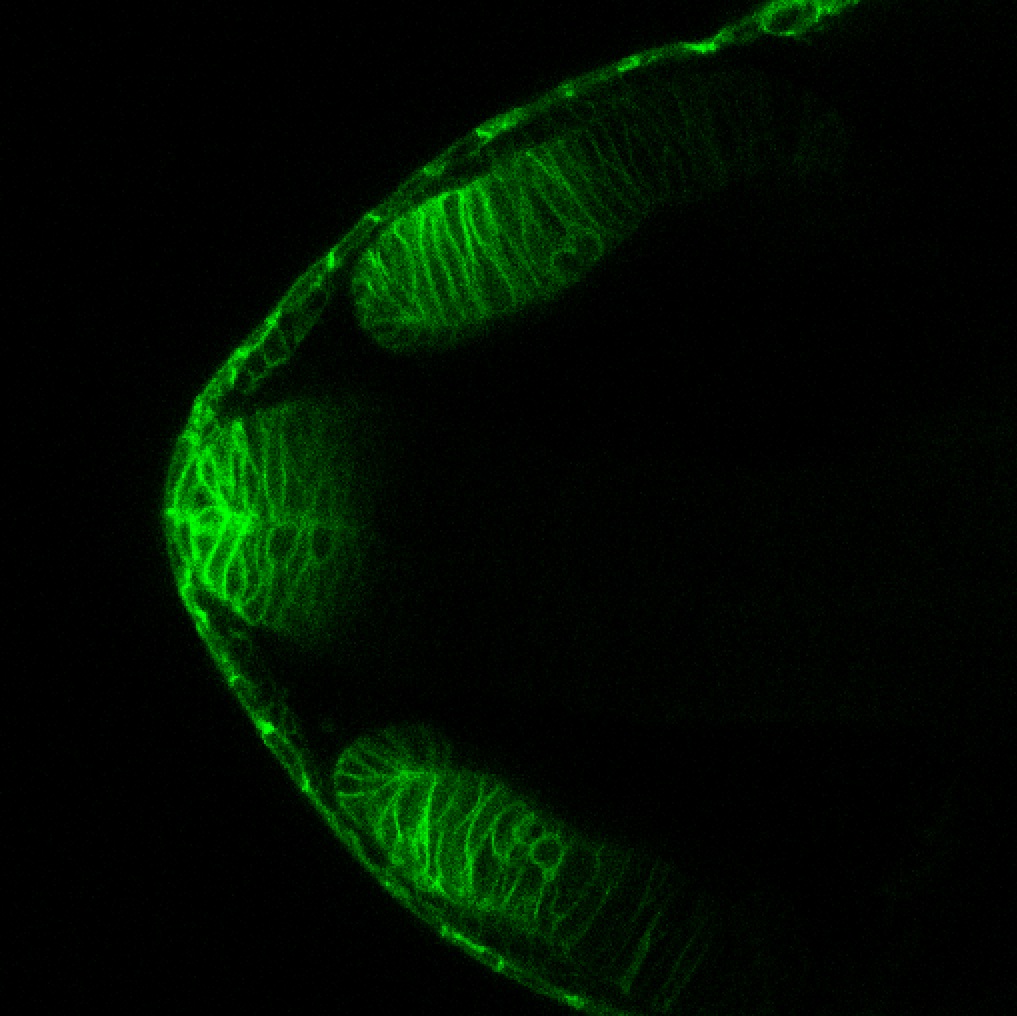
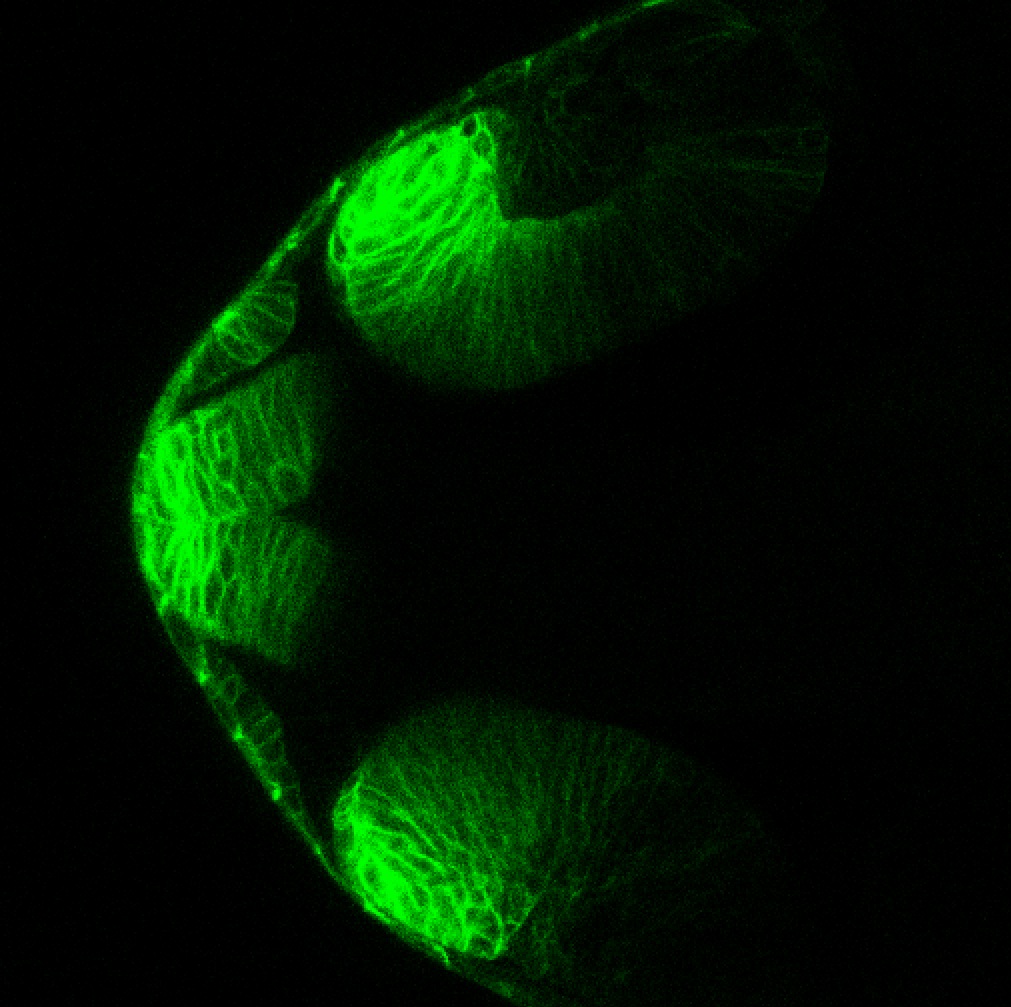
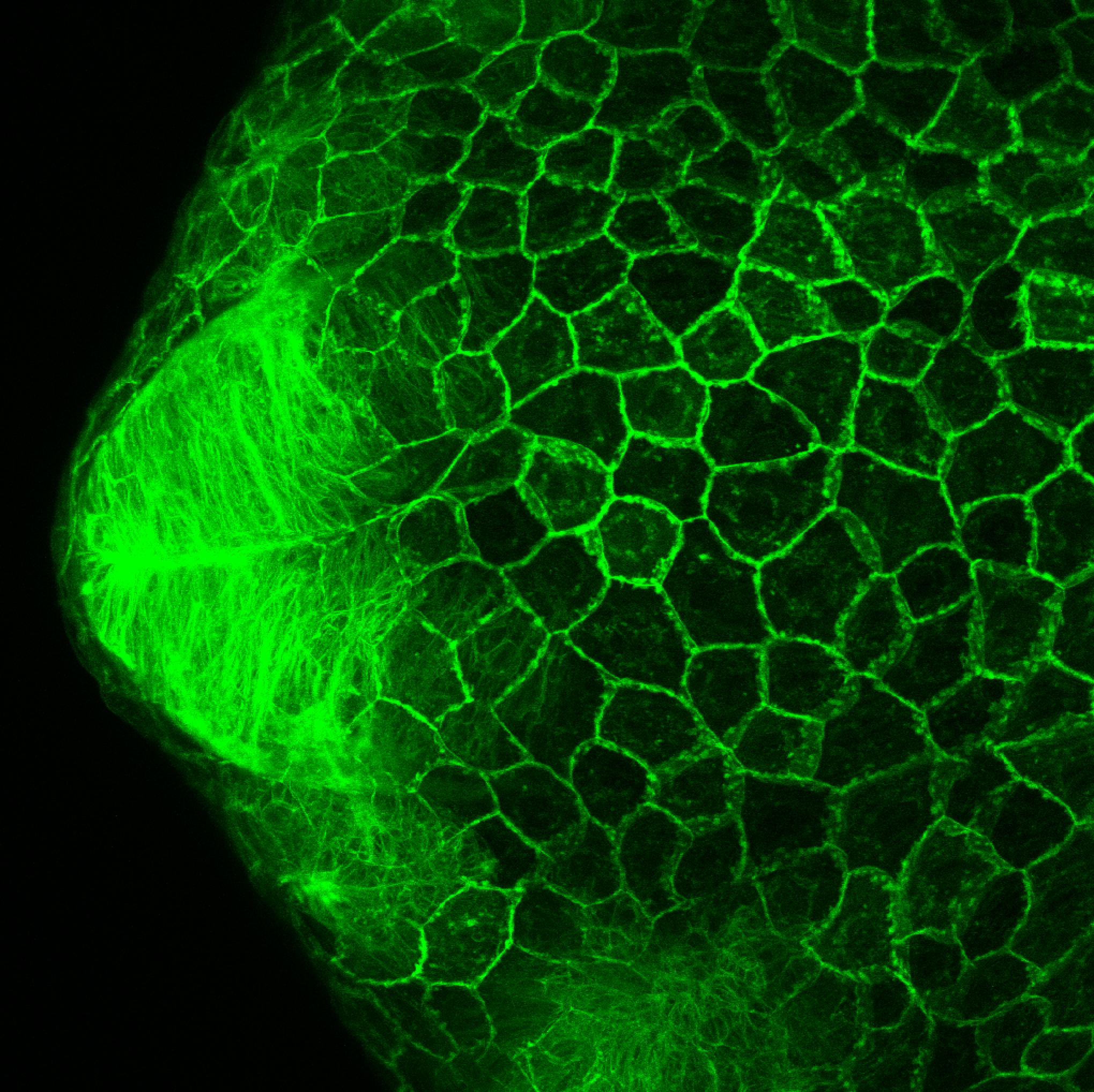
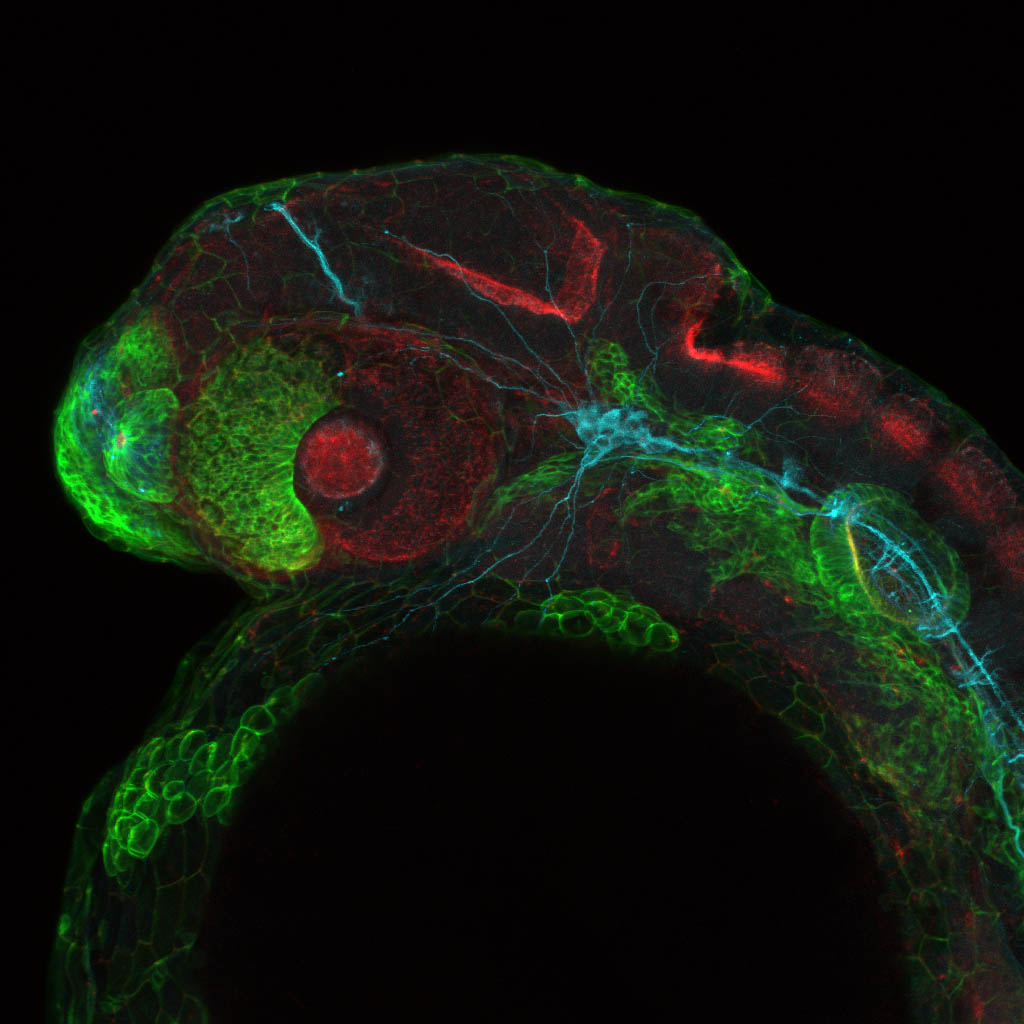
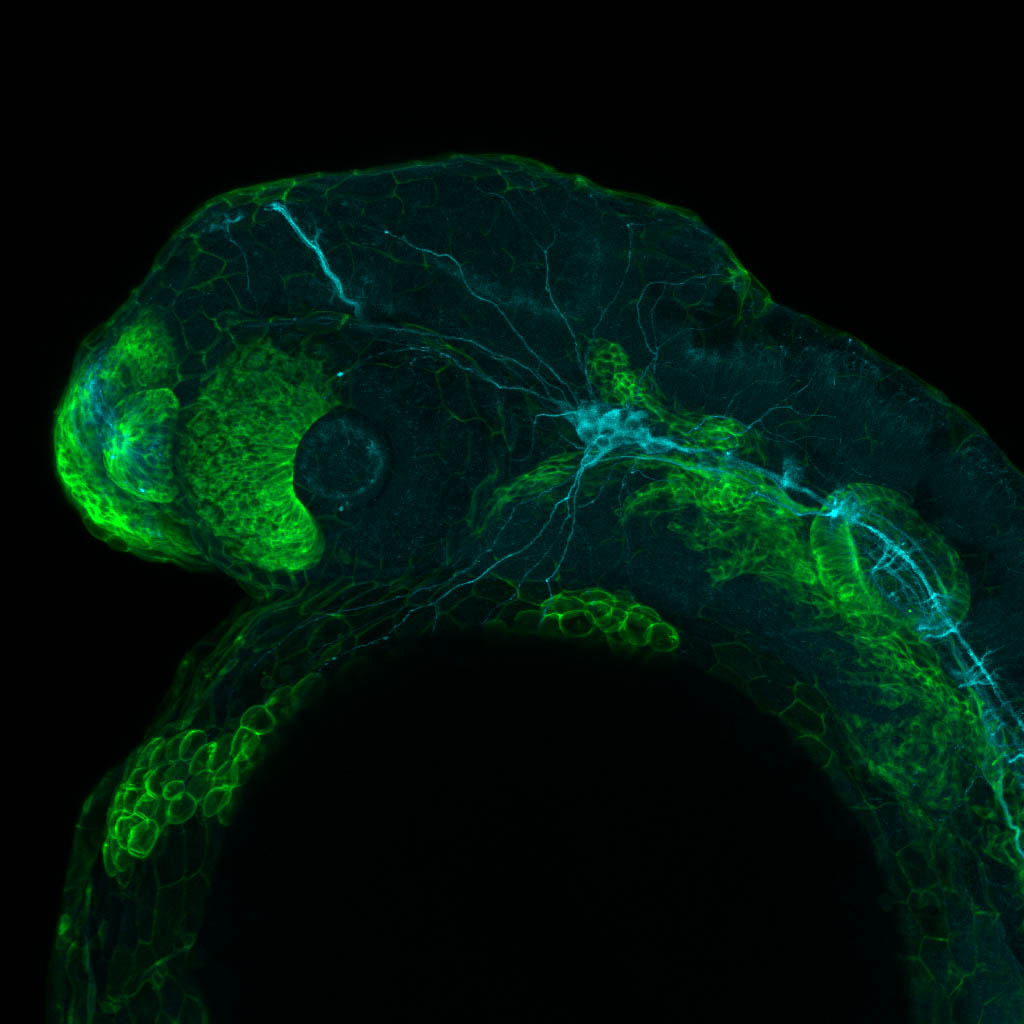
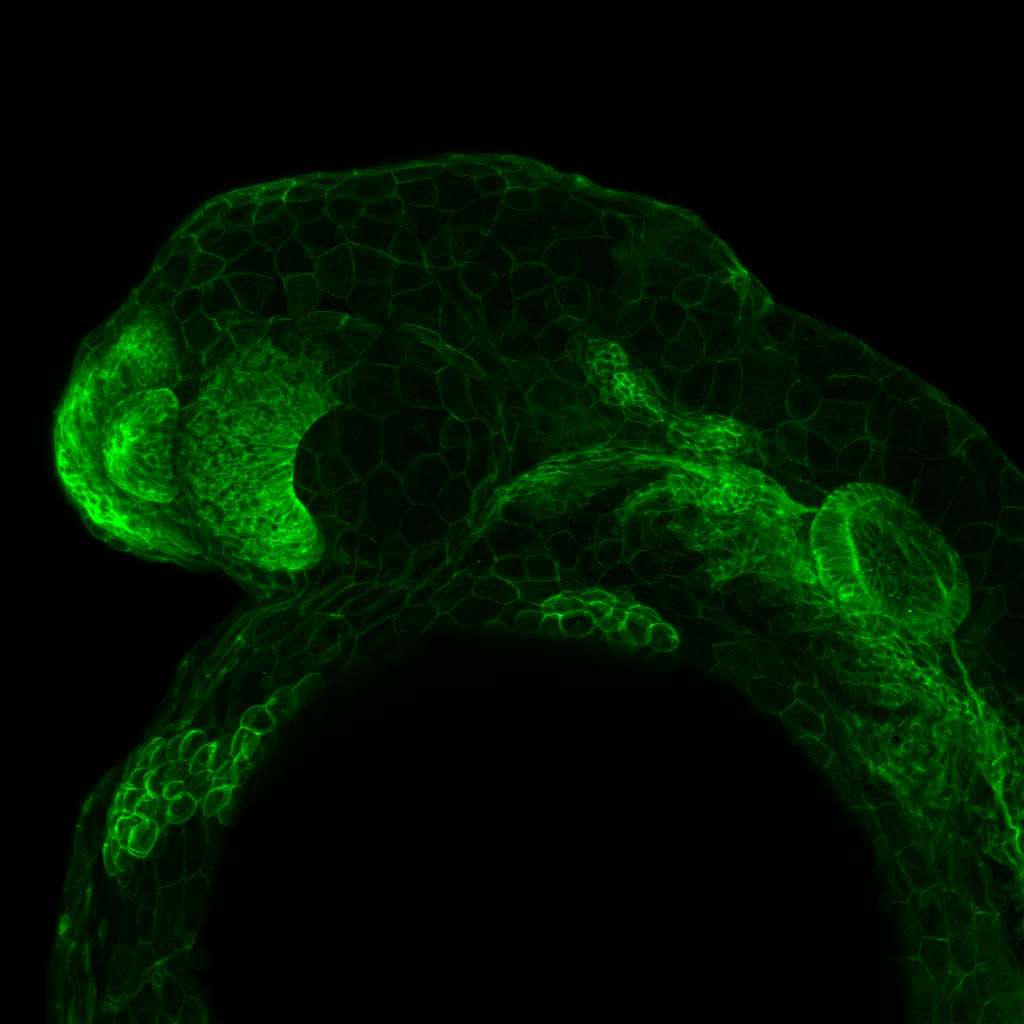
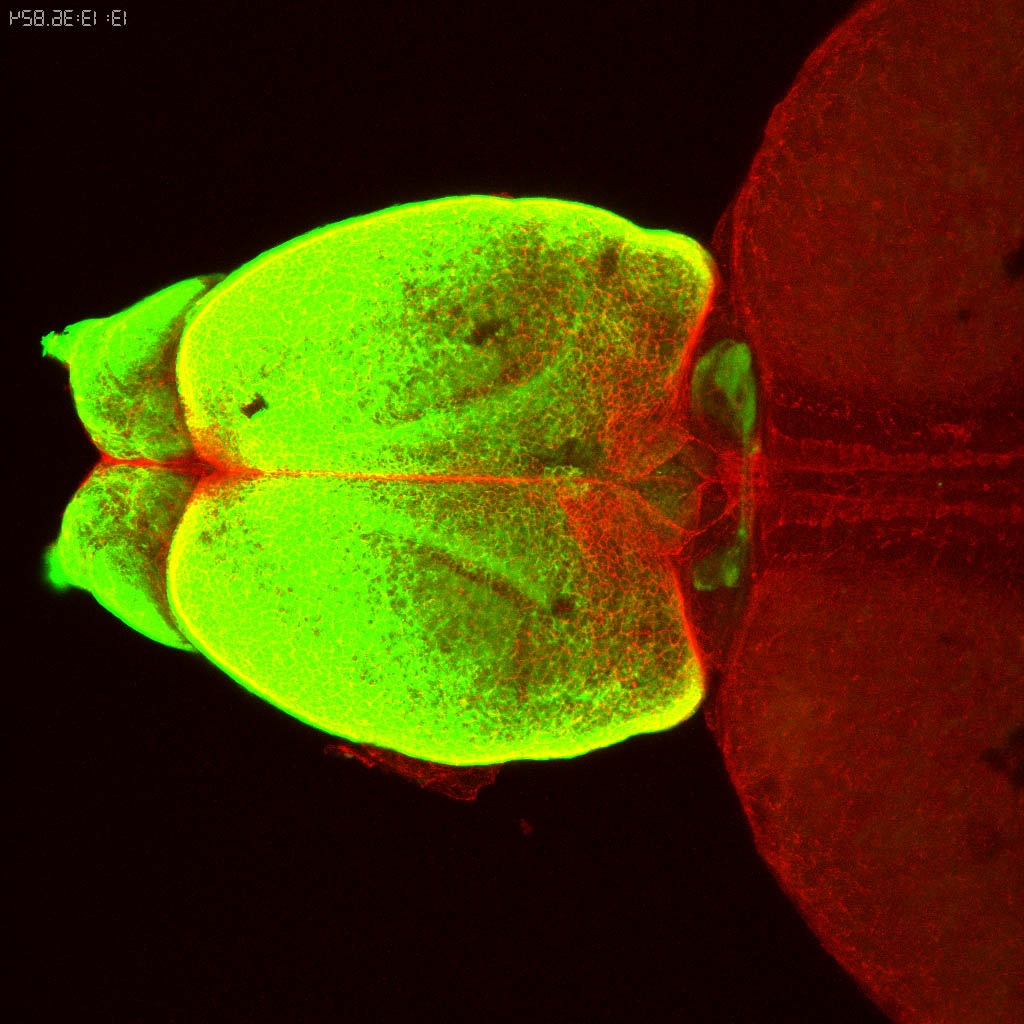
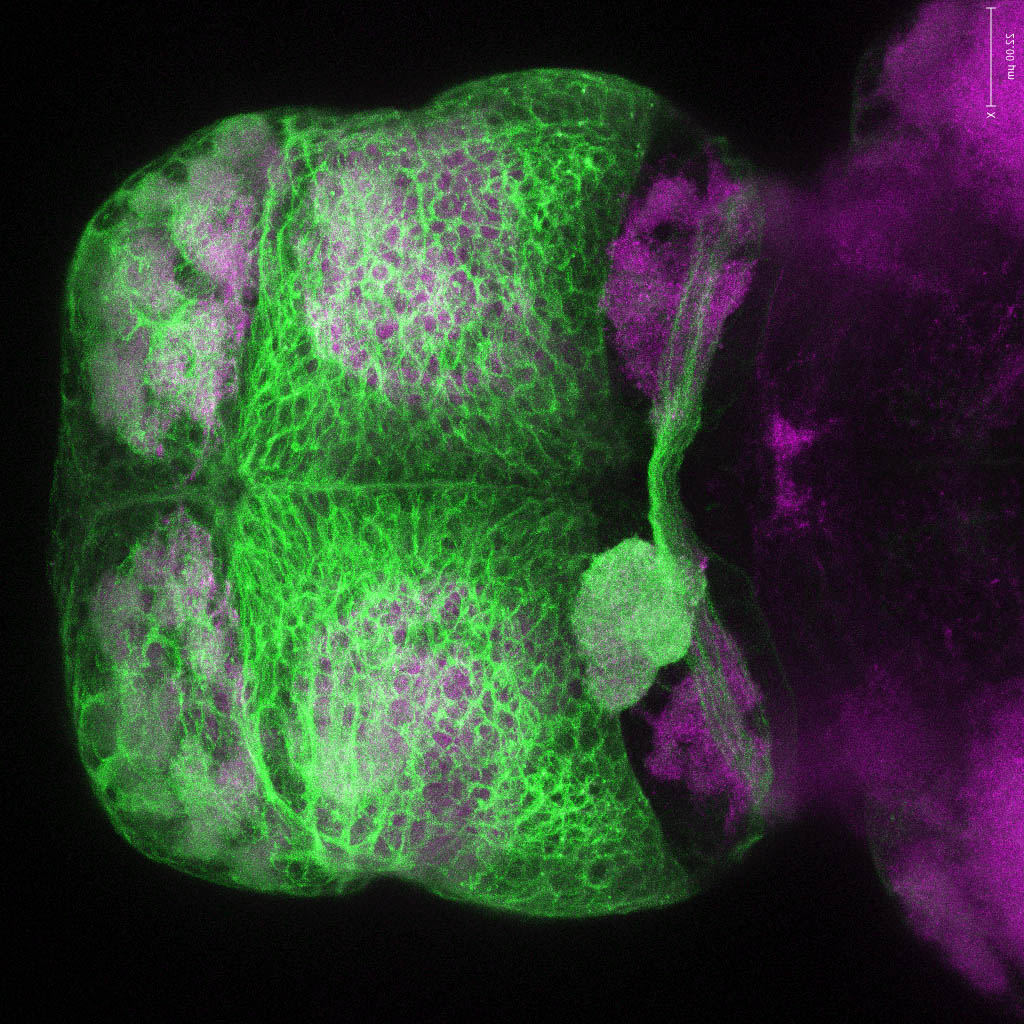
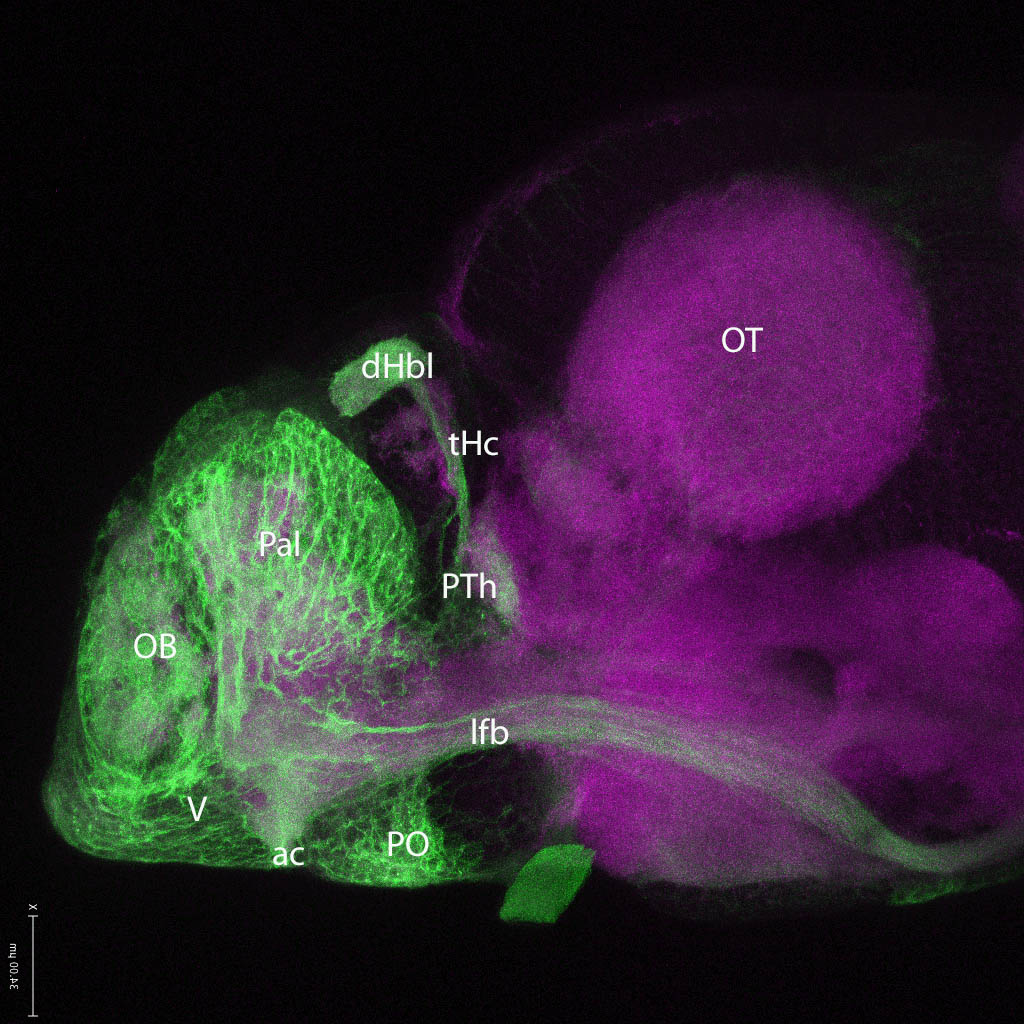
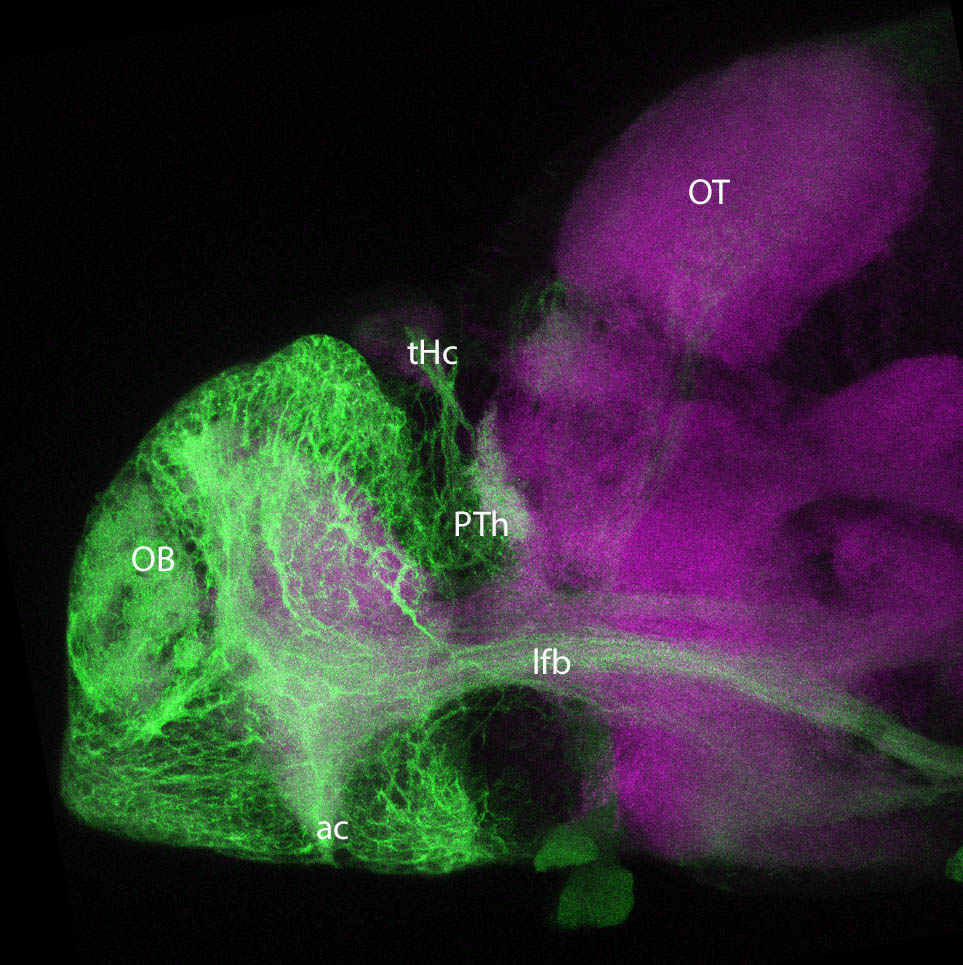
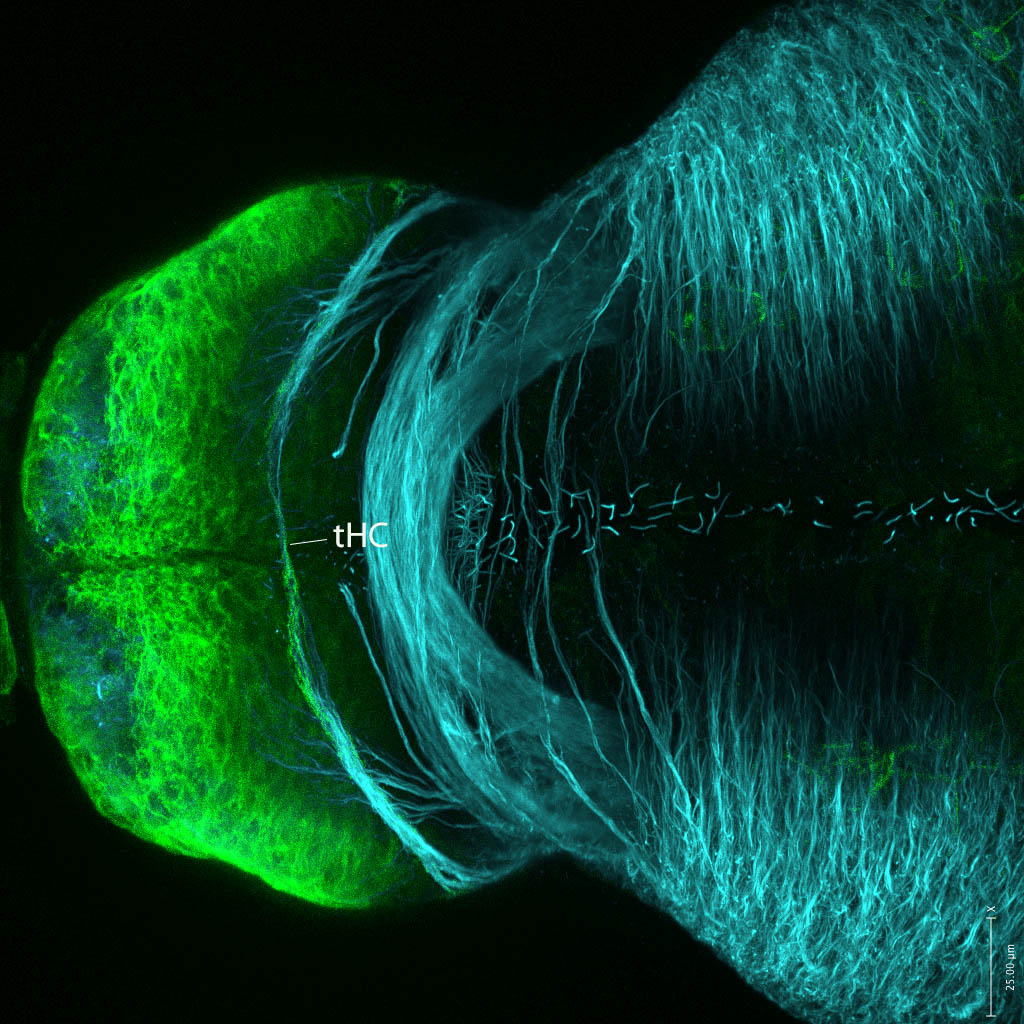
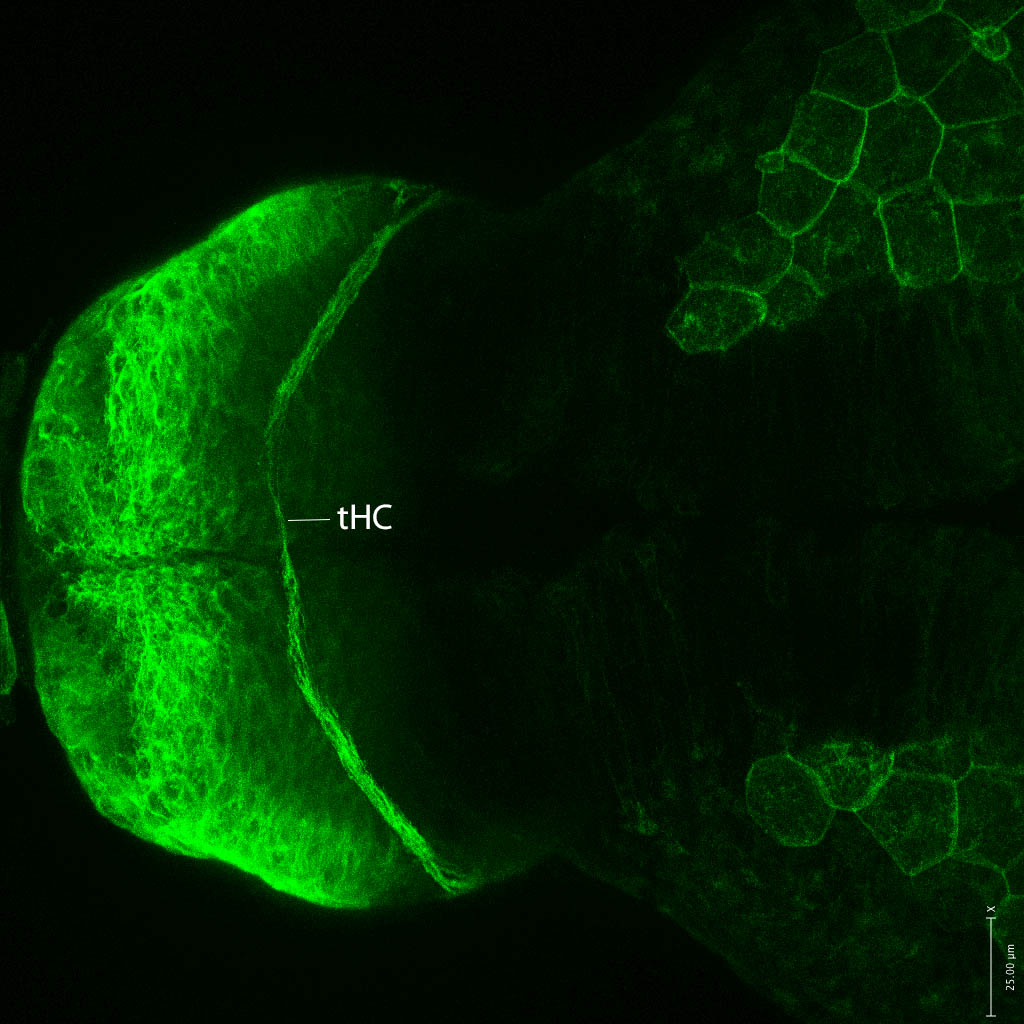
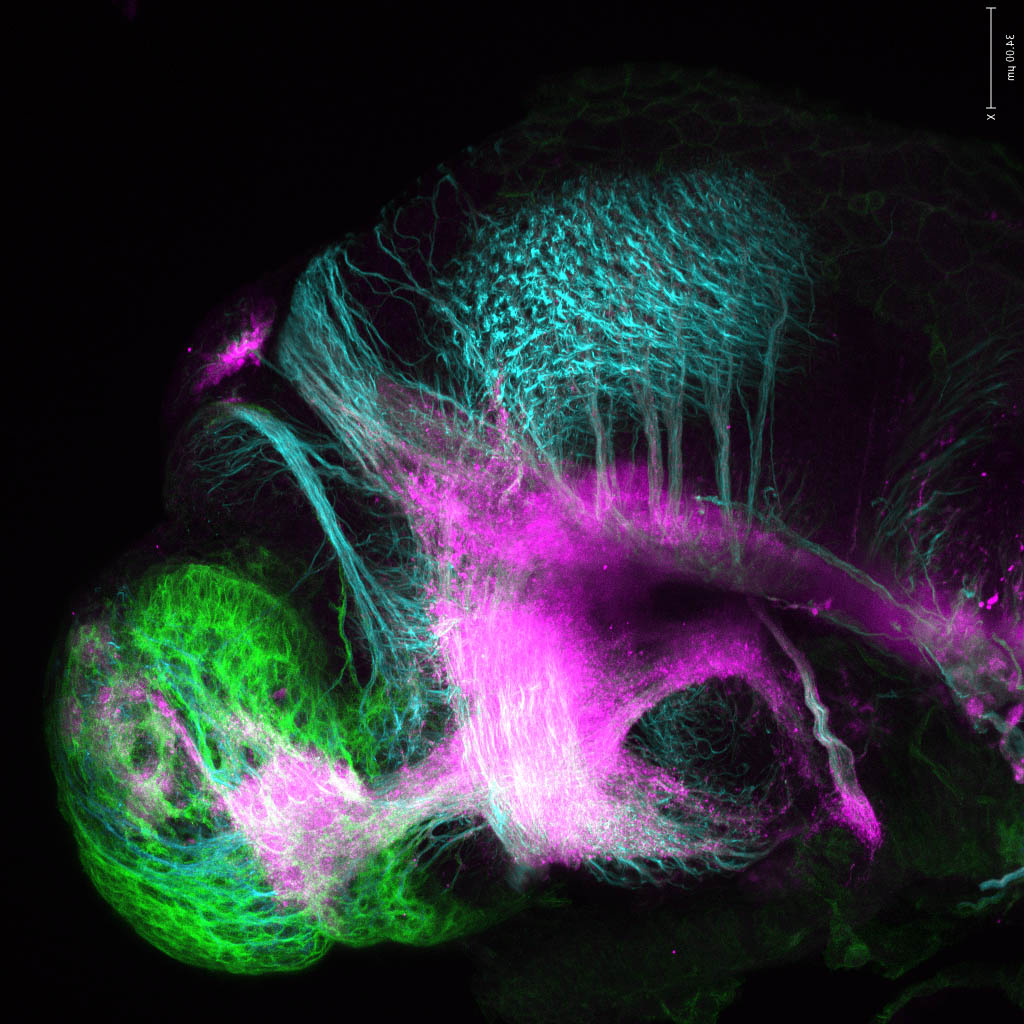
Expressed in:
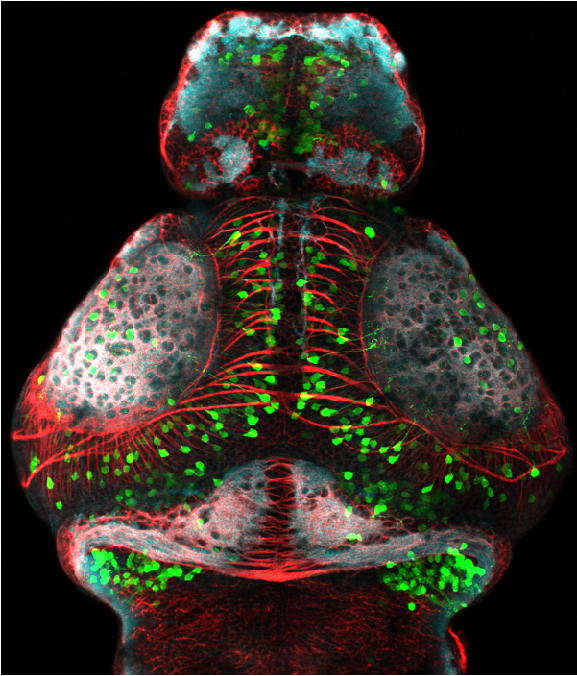
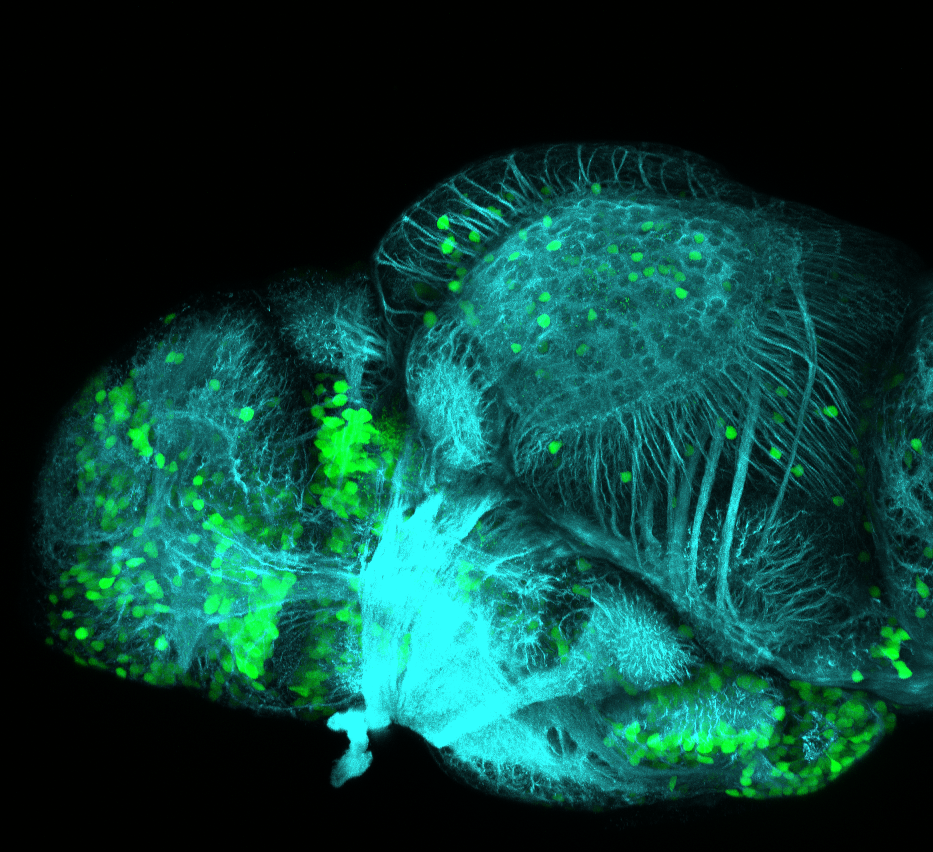
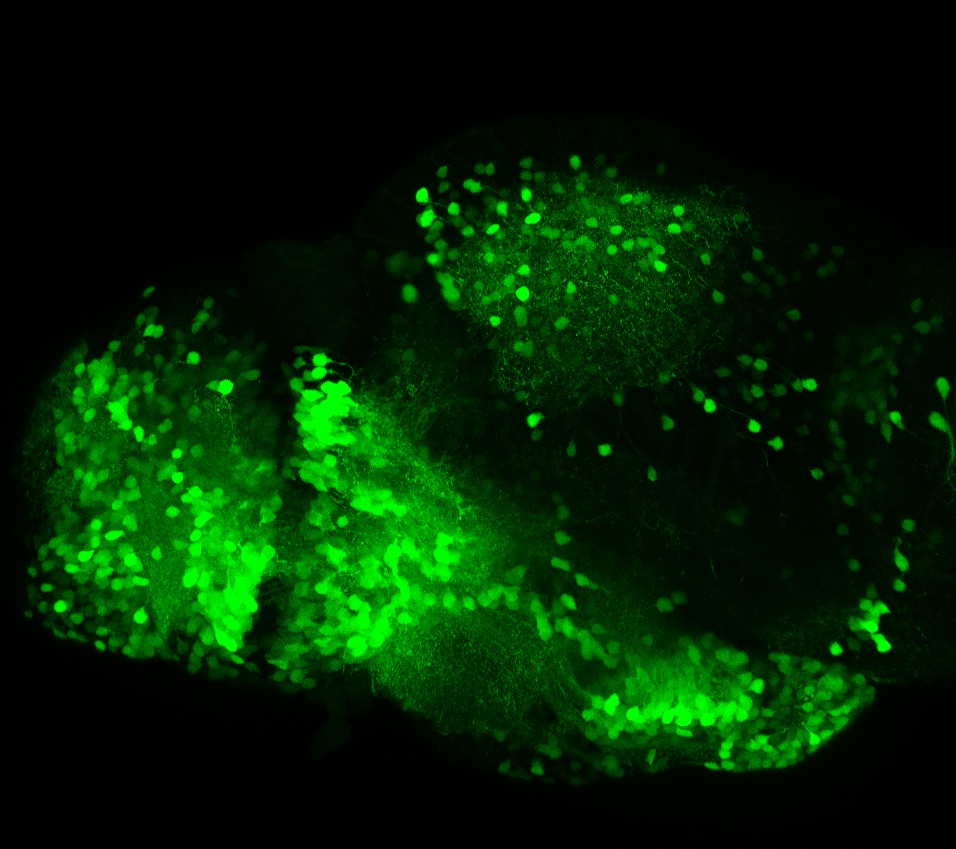
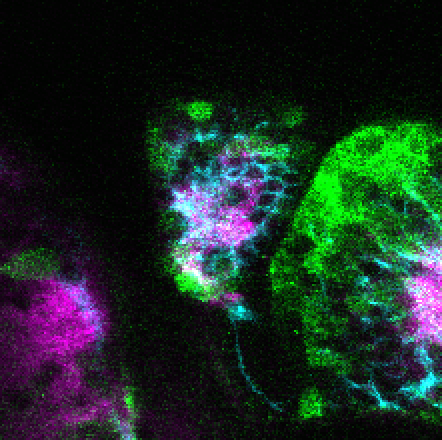
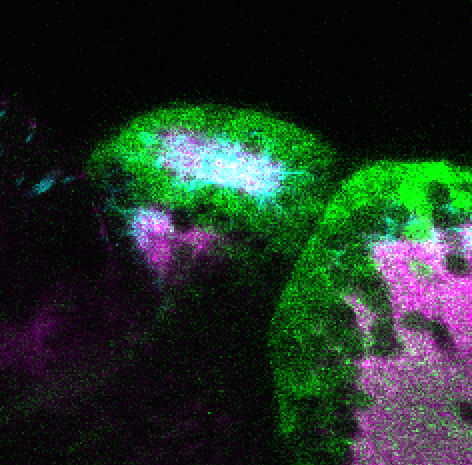
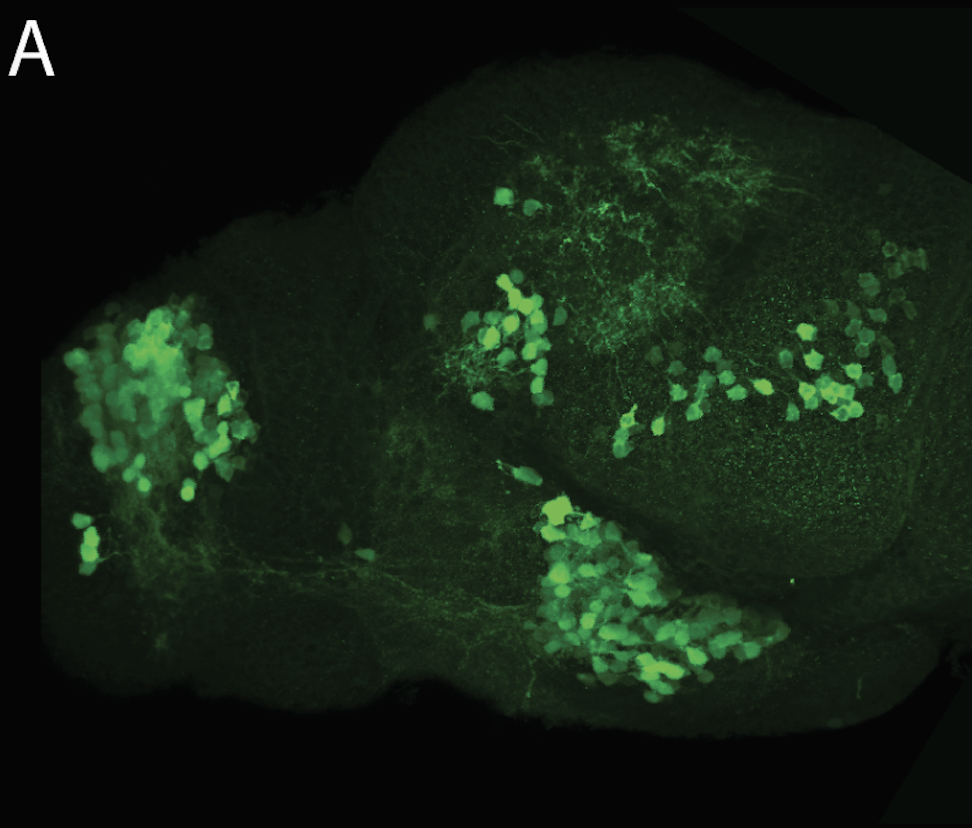
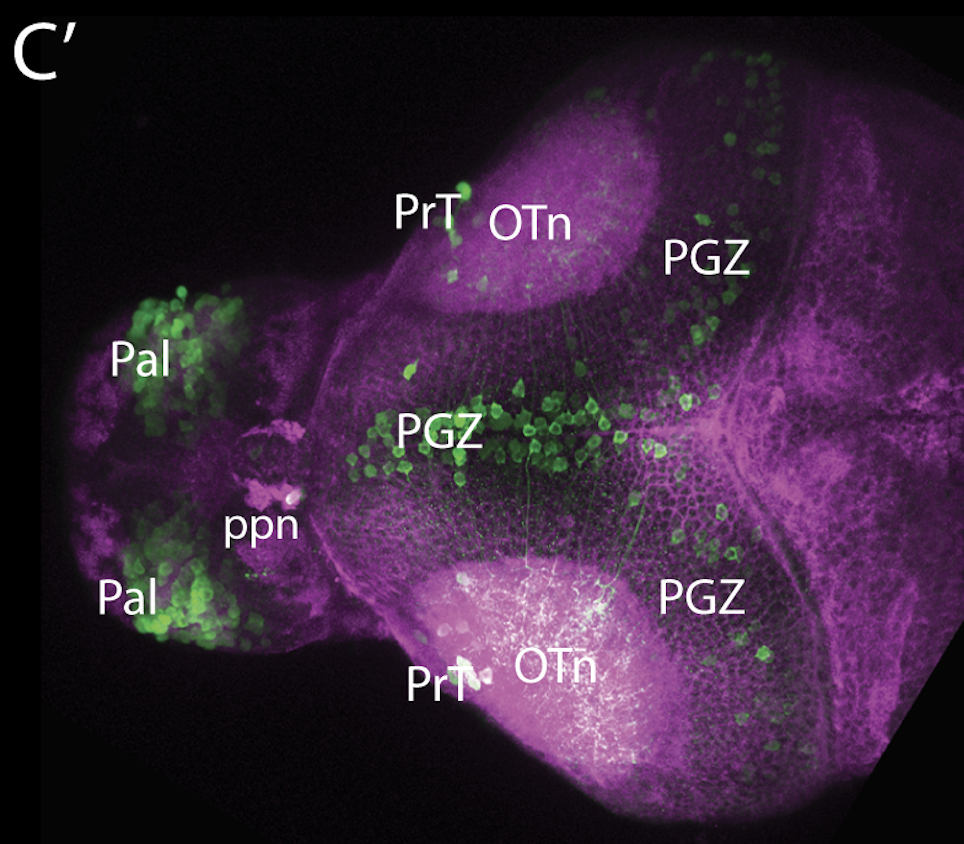
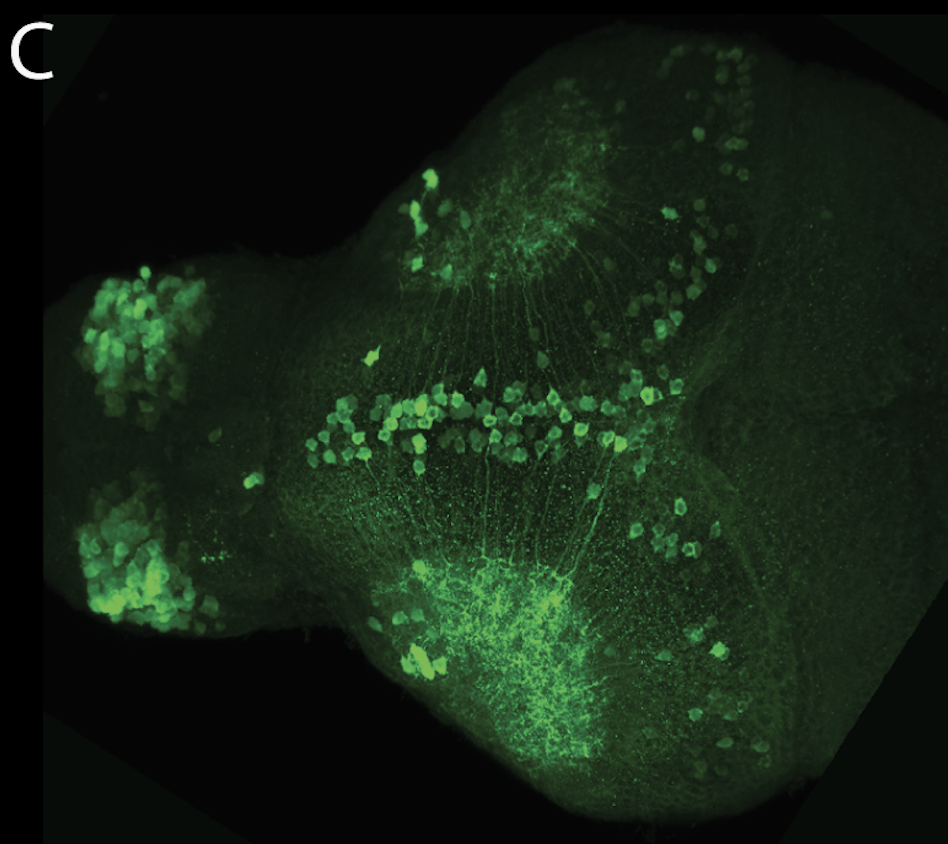
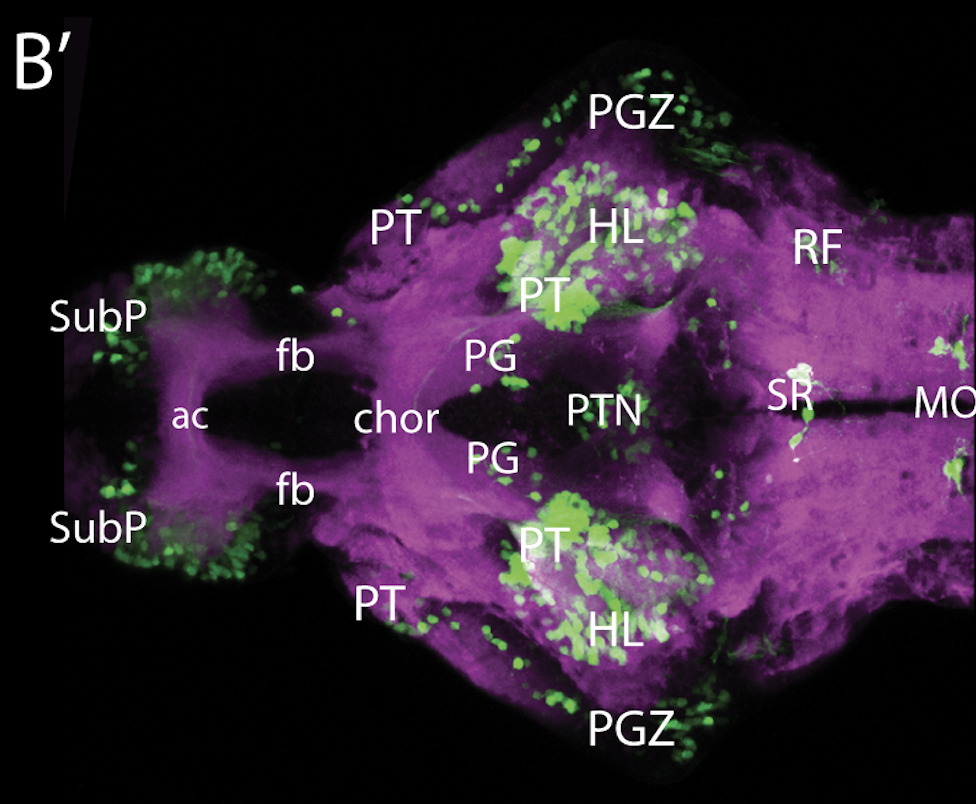
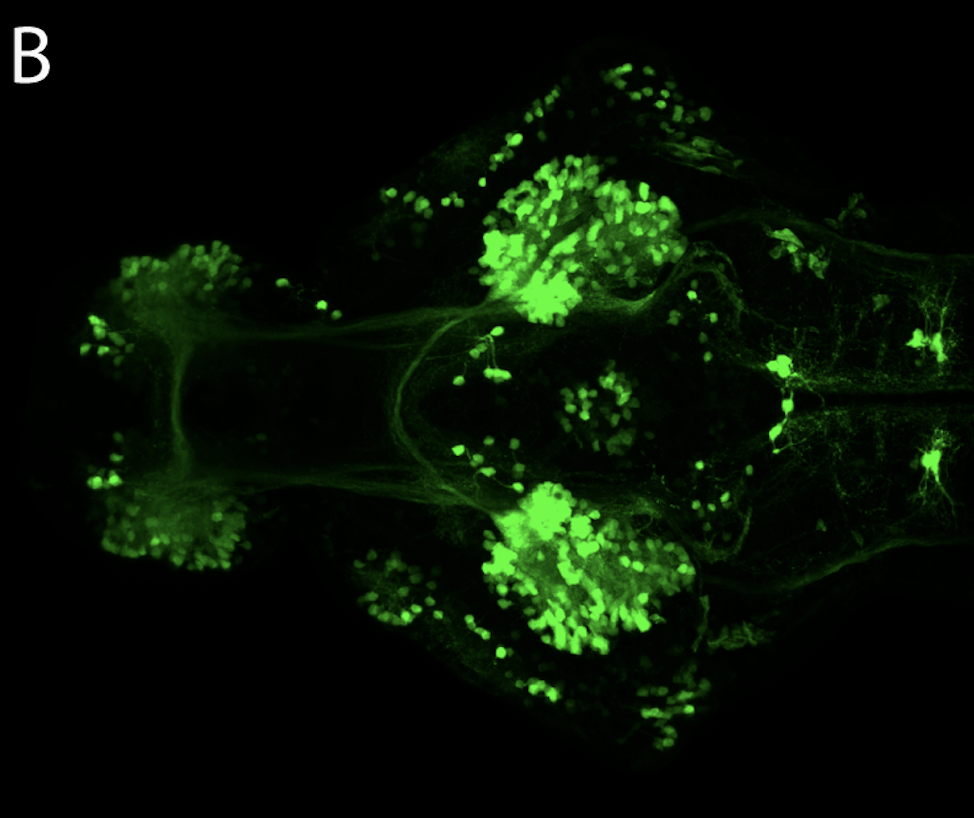
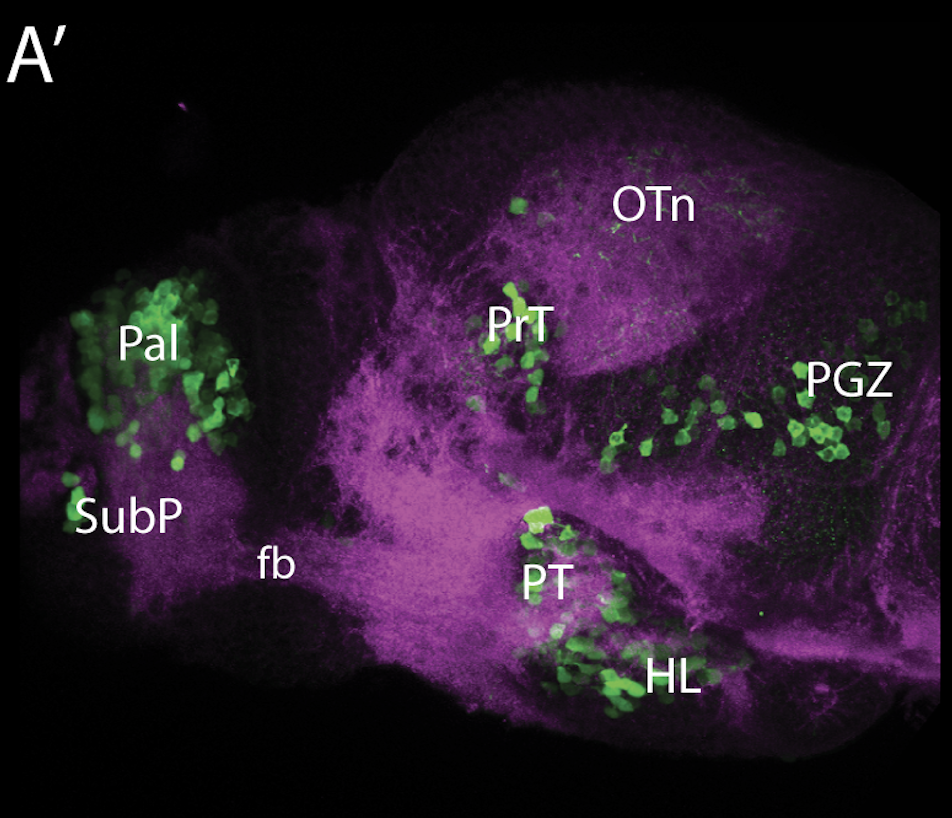
neuromasts, lateral line, olfactory epithelium, olfactory bulb, , pallium, subpallium, tract of the habenula commissure, nasal retina.
Key Publications
Haas, P., and Gilmour, D. (2006)
Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line.
Developmental Cell. 10(5):673-680.
Folgueira, M., Bayley, P., Navratilova, P., Becker, T.S., Wilson, S.W., and Clarke, J.D. (2012)
Morphogenesis underlying the development of the everted teleost telencephalon.
Neural Development. 7(1):32.
Valdivia, L.E., Young, R.M., Hawkins, T.A., Stickney, H.L., Cavodeassi, F., Schwarz, Q., Pullin, L.M., Villegas, R., Moro, E., Argenton, F., Allende, M.L., and Wilson, S.W. (2011)
Lef1-dependent Wnt/β-catenin signalling drives the proliferative engine that maintains tissue homeostasis during lateral line development.
Development (Cambridge, England). 138(18):3931-3941.
Valentin, G., Haas, P., and Gilmour, D. (2007)
The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b.
Current biology : CB. 17(12):1026-1031.
Picker, A., Cavodeassi, F., Machate, A., Bernauer, S., Hans, S., Abe, G., Kawakami, K., Wilson, S.W., and Brand, M. (2009)
Dynamic coupling of pattern formation and morphogenesis in the developing vertebrate retina.
PLoS Biology. 7(10):e1000214.















![[Improvision Data]ImageName=TimeStampMicroSeconds=3319455431398154TimeStamp=14:57:11.398 on 09 Mar 2009ChannelName=ChannelNo=1TimepointName=1TimepointNo=1ZPlane=1BlackPoint=0WhitePoint=255WhiteColour=255,255,255XCalibrationMicrons=1YCalibrationMicro](https://images.squarespace-cdn.com/content/v1/58065fb61b631b37ff3ce66a/1568631946526-34AZDYGG28NB7Q4A9WXX/Snapshot+of+Series008+%28vmat_tub_sv2_5d%29_10.jpg)
![[Improvision Data]ImageName=TimeStampMicroSeconds=3319455437197463TimeStamp=14:57:17.197 on 09 Mar 2009ChannelName=ChannelNo=1TimepointName=1TimepointNo=1ZPlane=1BlackPoint=0WhitePoint=255WhiteColour=255,255,255XCalibrationMicrons=1YCalibrationMicro](https://images.squarespace-cdn.com/content/v1/58065fb61b631b37ff3ce66a/1568631420219-DNOWMW8T30HIN5ZAE238/Snapshot+of+Series008+%28vmat_tub_sv2_5d%29_11.jpg)